Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103844 |
| Name | oriT_USDA1157|psymA |
| Organism | Sinorhizobium meliloti strain USDA1157 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP021794 (41612..41640 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_USDA1157|psymA
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file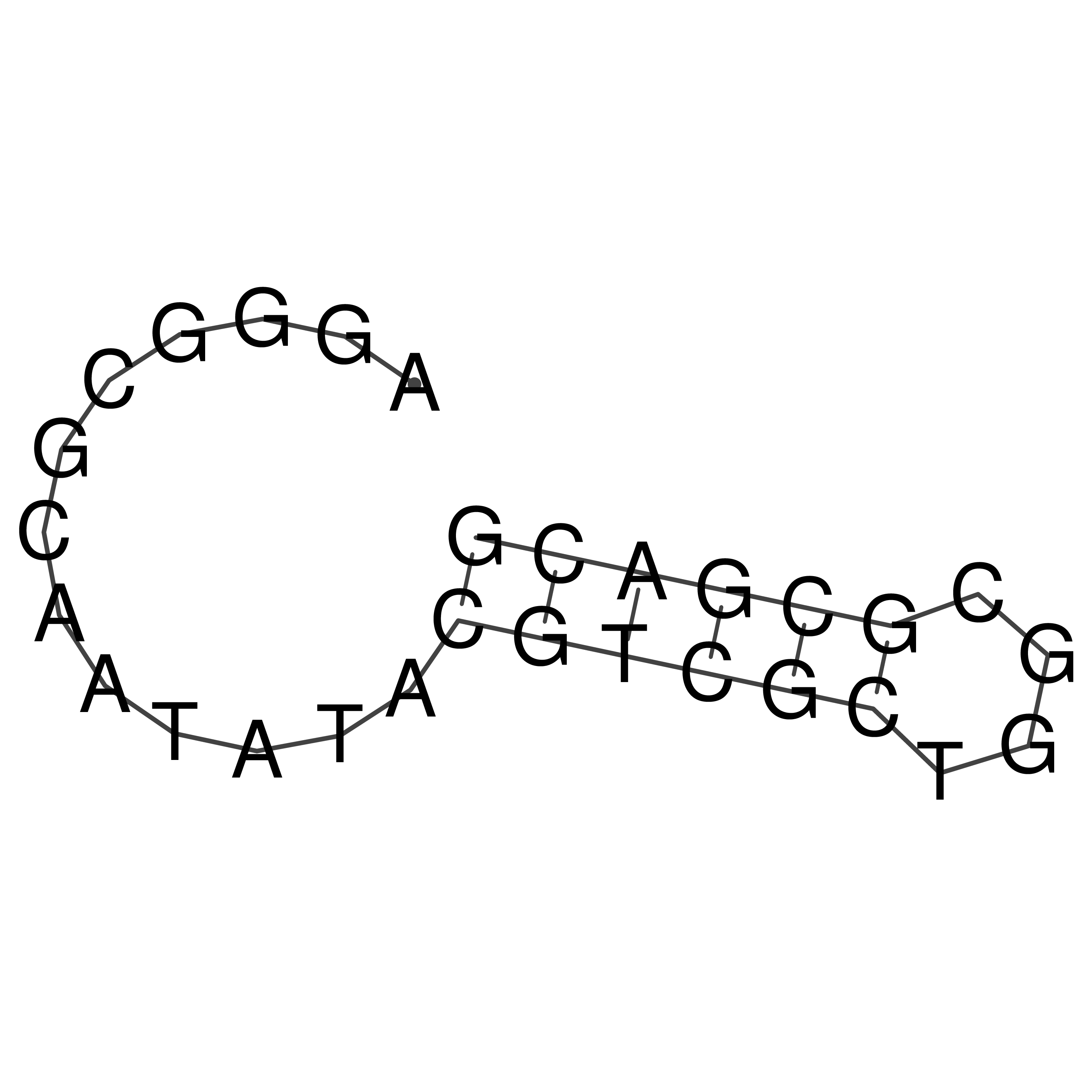
Relaxase
| ID | 2854 | GenBank | WP_088194634 |
| Name | traA_CDO31_RS19280_USDA1157|psymA |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 169864.47 Da Isoelectric Point: 9.2615
>WP_088194634.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAETGEAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRKQLNRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQHGRIVETETRAEIVDRIVADWGNARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRQVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDEQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFAALVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKRELVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVRKQGGDDAYRTAFAY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLARERREKLPRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVTNDPAAYGALRGSDRIMDKLLAAGRERKDALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSQEIISERQRRVIDRARSVTR
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAETGEAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRKQLNRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQHGRIVETETRAEIVDRIVADWGNARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRQVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDEQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFAALVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKRELVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVRKQGGDDAYRTAFAY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLARERREKLPRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVTNDPAAYGALRGSDRIMDKLLAAGRERKDALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSQEIISERQRRVIDRARSVTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2685 | GenBank | WP_014528597 |
| Name | traG_CDO31_RS19295_USDA1157|psymA |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70213.22 Da Isoelectric Point: 9.3518
>WP_014528597.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium meliloti]
MALKAKPHPSLLVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRKPVAMASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFDPNDPATWGQGGKVP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSIGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIEADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARSISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAVP
MALKAKPHPSLLVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRKPVAMASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFDPNDPATWGQGGKVP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSIGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIEADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARSISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1374514..1385398
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CDO31_RS26525 (CDO31_26530) | 1371397..1372244 | + | 848 | Protein_1364 | alpha/beta fold hydrolase | - |
| CDO31_RS26530 (CDO31_26535) | 1372294..1372941 | + | 648 | WP_010967700 | dihydrofolate reductase family protein | - |
| CDO31_RS26535 (CDO31_26540) | 1373015..1373266 | + | 252 | WP_010967699 | hypothetical protein | - |
| CDO31_RS26540 (CDO31_26545) | 1373527..1373895 | - | 369 | WP_010967698 | transcriptional regulator | - |
| CDO31_RS26545 (CDO31_26550) | 1373992..1374510 | + | 519 | WP_017277528 | hypothetical protein | - |
| CDO31_RS26550 (CDO31_26555) | 1374514..1375185 | + | 672 | WP_010967696 | lytic transglycosylase domain-containing protein | virB1 |
| CDO31_RS26555 (CDO31_26560) | 1375182..1375481 | + | 300 | WP_003526741 | TrbC/VirB2 family protein | virB2 |
| CDO31_RS26560 (CDO31_26565) | 1375486..1375827 | + | 342 | WP_003526740 | type IV secretion system protein VirB3 | virB3 |
| CDO31_RS26565 (CDO31_26570) | 1375802..1378198 | + | 2397 | WP_017266273 | VirB4 family type IV secretion system protein | virb4 |
| CDO31_RS26570 (CDO31_26575) | 1378198..1378899 | + | 702 | WP_014531544 | P-type DNA transfer protein VirB5 | virB5 |
| CDO31_RS26575 (CDO31_26580) | 1378896..1379111 | + | 216 | WP_014531545 | EexN family lipoprotein | - |
| CDO31_RS26580 (CDO31_26585) | 1379134..1379697 | + | 564 | Protein_1375 | type IV secretion system protein | - |
| CDO31_RS26585 (CDO31_26590) | 1379735..1380937 | - | 1203 | WP_010967204 | IS256-like element ISRm3 family transposase | - |
| CDO31_RS26595 (CDO31_26600) | 1380989..1381375 | + | 387 | Protein_1377 | type IV secretion system protein | - |
| CDO31_RS38615 (CDO31_26605) | 1381372..1381686 | + | 315 | WP_013845458 | hypothetical protein | - |
| CDO31_RS26605 (CDO31_26610) | 1381691..1382362 | + | 672 | WP_013845457 | virB8 family protein | virB8 |
| CDO31_RS26610 (CDO31_26615) | 1382362..1383216 | + | 855 | WP_014531548 | P-type conjugative transfer protein VirB9 | virB9 |
| CDO31_RS26615 (CDO31_26620) | 1383226..1384398 | + | 1173 | WP_088194881 | type IV secretion system protein VirB10 | virB10 |
| CDO31_RS26620 (CDO31_26625) | 1384409..1385398 | + | 990 | WP_088194882 | P-type DNA transfer ATPase VirB11 | virB11 |
| CDO31_RS26625 (CDO31_26630) | 1385709..1386935 | - | 1227 | WP_013845453 | MFS transporter | - |
| CDO31_RS26630 (CDO31_26635) | 1386972..1387583 | - | 612 | WP_014531550 | TetR/AcrR family transcriptional regulator | - |
| CDO31_RS26635 (CDO31_26640) | 1388110..1388295 | + | 186 | WP_013845451 | periplasmic nitrate reductase, NapE protein | - |
| CDO31_RS26640 (CDO31_26645) | 1388295..1388795 | + | 501 | WP_088194883 | ferredoxin-type protein NapF | - |
| CDO31_RS26645 (CDO31_26650) | 1388788..1389075 | + | 288 | WP_088194884 | chaperone NapD | - |
Host bacterium
| ID | 4284 | GenBank | NZ_CP021794 |
| Plasmid name | USDA1157|psymA | Incompatibility group | - |
| Plasmid size | 1433621 bp | Coordinate of oriT [Strand] | 41612..41640 [-] |
| Host baterium | Sinorhizobium meliloti strain USDA1157 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | ncrA |
| Degradation gene | - |
| Symbiosis gene | nodM, nifN, nodD, nodA, nodB, nodC, nodI, nodJ, nodE, nodF, nodH, nifX, nifE, nifK, nifD, nifH, fixA, fixB, fixC, fixX, nifB, fixU, fixO, fixN, fixG, fixS |
| Anti-CRISPR | AcrIIA7 |