Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103816 |
| Name | oriT_pUPC1 |
| Organism | Enterobacter cancerogenus strain UPC1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LR881937 (81346..81458 [-], 113 nt) |
| oriT length | 113 nt |
| IRs (inverted repeats) | 93..99, 103..109 (TAATTTA..TAAATTA) 29..34, 41..46 (GCAAAA..TTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 113 nt
>oriT_pUPC1
TTCTCACCACAAAAGCACCACACCCCACGCAAAACGTACTTTTTGCTGATTGGCTATTCAGTTCAGTAACTTAGCGTTTGAATAATGTGTAATAATTTATTTTAAATTACTAA
TTCTCACCACAAAAGCACCACACCCCACGCAAAACGTACTTTTTGCTGATTGGCTATTCAGTTCAGTAACTTAGCGTTTGAATAATGTGTAATAATTTATTTTAAATTACTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file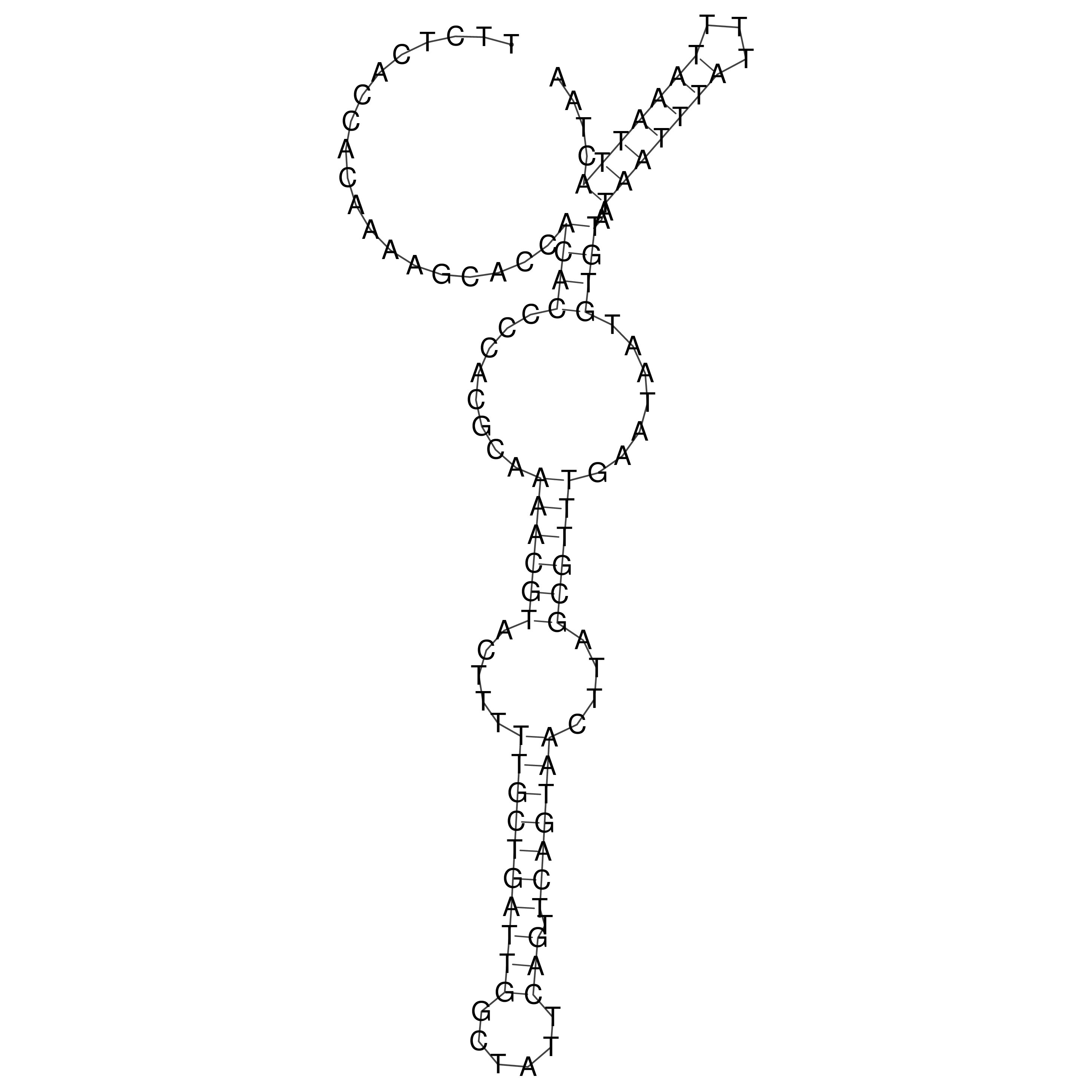
Relaxase
| ID | 2832 | GenBank | WP_202562368 |
| Name | traI_ENTER_RS22910_pUPC1 |
UniProt ID | _ |
| Length | 1745 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1745 a.a. Molecular weight: 188888.87 Da Isoelectric Point: 6.3484
>WP_202562368.1 conjugative transfer relaxase/helicase TraI [Enterobacter cancerogenus]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGEGAKALGLSGSVDVKTFTRVLEGKLPDGSDLSRAQ
DGTNKHRPGYDLTFSAPKSISVMAMLGGDTRLIDAHNRAVDTAIKQIEALASTRVMTDGKSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHDGTWRTLSSDKIGKTGFIENVYANQIAFGRLYRAALKDDVTA
MGYETETVGKHGMWELKGVPTEPYSSRTKAIREAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMAT
LKETGFDIQAYREAADQRVREGAVPVTTPESADAPAAVSQSIAILSDRRARFTYSELLATTLGQLPARPG
MVEQAREGIDAAIKNEQLIPLDKEKGLFTSNVHVLDELSVSALSRELQQQGRVDVFPDKSVPRSRAFGDA
VSVLAQDRSPVAIISGQGGAAGQRERVAELAMMAREQGRDVQIMAADRRSAANLARDERLSGELITDRRG
LTEGMTFIPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRIGTGSALTVMKEAGVSTLAWQG
GEKTRVSVISEADRRERYDRLAADFAKSIRAGEEAVVQISGAREQAALAATVRDALKGEKVLGERDISIT
TLEPAWLDGRHRQVRDHYREGMVMERWNAEERSRERFVIDRVTARNNSLTLRNAQGETQVTRLTELDSSW
SLYRTGTLQVTEGDRLAVLGQTQGARLKGGDSVTVTGIDEKGIHVSLPGRKTQVVLPAGDSPFTAPKIGQ
GWVESPGRSVSDSATVYASLTQREMDNATLNGLARSGAEIRLYSAQTVQKTEEKLSRQSAWTVVTAQVKE
AAGKDSLGEALAHQKAALHTPEQQAIHLAIPSLEGNGLAFTRPQLMAAAKDFAQDKLSLAAIEDEIARQT
RSGALISVPVSQGNGLQQLVSRQSYNAEKSIIRHVLEGKDAVTPLMGKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEEQRPRVIGVAPTHRAVGEMRDAGVPESQTLAAFIHDTQ
QQLRGGERPDFSNVLFLIDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DTAVMQEIVRQTPALRPAVYSLIERNIGSALTTLESVAPQQVPRRPDAWQPDGSVMEFSREHEKAIADAI
KAGDLAPGGQPATLLEAIVKDYTGRTPQAQTIVITALNADRRQLNAMIHDARREAGELGEKEAIVPVLTP
ANIRDGELRRMDTWQTHASSMVLLDNTYYSIDALDKDAHLVTLKDAQGNTRSLSPAQAATEGVTLYRQDT
IAVSQGDRMRFSKSDNERGFVANSVWTVSDIKGDIVTLTDGRQTSTVNPSAERAEQHIDLAYAVTAHGAQ
GASEPFSISLQGTDGGRKQMVNFESAYVALSRMKQHAQVYTDNREKWVAAMEKSQAKSTAHDIVEPRNDR
AVANAARLTATAKALGEVPAGRAALRQAGLLAESTMAKFISPGRKYPQPHVALPAFDRNGRQAGVWLSTL
TSGDGQLQGLGSEGRVMGSEDAAFAGLQASRNGESLMARDMAEGVRLARENPQSGVVVRIGDAEGRPWNP
GAITGGRIWADGVPDSTGTQHGEKIPPEVLAQQALEAQQRRELEKRAEDAVRELARGSEKAEDAAGAVRE
LARELSEDKTRERAADVTLPESPDVRARNEAVTRVAHENVQRDRLQQMERDTVRVLEREKTMGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGEGAKALGLSGSVDVKTFTRVLEGKLPDGSDLSRAQ
DGTNKHRPGYDLTFSAPKSISVMAMLGGDTRLIDAHNRAVDTAIKQIEALASTRVMTDGKSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHDGTWRTLSSDKIGKTGFIENVYANQIAFGRLYRAALKDDVTA
MGYETETVGKHGMWELKGVPTEPYSSRTKAIREAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMAT
LKETGFDIQAYREAADQRVREGAVPVTTPESADAPAAVSQSIAILSDRRARFTYSELLATTLGQLPARPG
MVEQAREGIDAAIKNEQLIPLDKEKGLFTSNVHVLDELSVSALSRELQQQGRVDVFPDKSVPRSRAFGDA
VSVLAQDRSPVAIISGQGGAAGQRERVAELAMMAREQGRDVQIMAADRRSAANLARDERLSGELITDRRG
LTEGMTFIPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRIGTGSALTVMKEAGVSTLAWQG
GEKTRVSVISEADRRERYDRLAADFAKSIRAGEEAVVQISGAREQAALAATVRDALKGEKVLGERDISIT
TLEPAWLDGRHRQVRDHYREGMVMERWNAEERSRERFVIDRVTARNNSLTLRNAQGETQVTRLTELDSSW
SLYRTGTLQVTEGDRLAVLGQTQGARLKGGDSVTVTGIDEKGIHVSLPGRKTQVVLPAGDSPFTAPKIGQ
GWVESPGRSVSDSATVYASLTQREMDNATLNGLARSGAEIRLYSAQTVQKTEEKLSRQSAWTVVTAQVKE
AAGKDSLGEALAHQKAALHTPEQQAIHLAIPSLEGNGLAFTRPQLMAAAKDFAQDKLSLAAIEDEIARQT
RSGALISVPVSQGNGLQQLVSRQSYNAEKSIIRHVLEGKDAVTPLMGKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEEQRPRVIGVAPTHRAVGEMRDAGVPESQTLAAFIHDTQ
QQLRGGERPDFSNVLFLIDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DTAVMQEIVRQTPALRPAVYSLIERNIGSALTTLESVAPQQVPRRPDAWQPDGSVMEFSREHEKAIADAI
KAGDLAPGGQPATLLEAIVKDYTGRTPQAQTIVITALNADRRQLNAMIHDARREAGELGEKEAIVPVLTP
ANIRDGELRRMDTWQTHASSMVLLDNTYYSIDALDKDAHLVTLKDAQGNTRSLSPAQAATEGVTLYRQDT
IAVSQGDRMRFSKSDNERGFVANSVWTVSDIKGDIVTLTDGRQTSTVNPSAERAEQHIDLAYAVTAHGAQ
GASEPFSISLQGTDGGRKQMVNFESAYVALSRMKQHAQVYTDNREKWVAAMEKSQAKSTAHDIVEPRNDR
AVANAARLTATAKALGEVPAGRAALRQAGLLAESTMAKFISPGRKYPQPHVALPAFDRNGRQAGVWLSTL
TSGDGQLQGLGSEGRVMGSEDAAFAGLQASRNGESLMARDMAEGVRLARENPQSGVVVRIGDAEGRPWNP
GAITGGRIWADGVPDSTGTQHGEKIPPEVLAQQALEAQQRRELEKRAEDAVRELARGSEKAEDAAGAVRE
LARELSEDKTRERAADVTLPESPDVRARNEAVTRVAHENVQRDRLQQMERDTVRVLEREKTMGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2658 | GenBank | WP_202562370 |
| Name | traD_ENTER_RS22915_pUPC1 |
UniProt ID | _ |
| Length | 755 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 755 a.a. Molecular weight: 84886.70 Da Isoelectric Point: 5.0333
>WP_202562370.1 type IV conjugative transfer system coupling protein TraD [Enterobacter cancerogenus]
MSFNAKDMTQGGQIASMRIRMFSQIANIILYCLVILFGIIVALTLAVKLSWQTFVNGMAWYWCQTLAPMR
DIFRSQPVYTIRYYGKELKYNAQQLLQDKYVIWCADQVWTAFILAAIIAAVVCVIAFFVTTWILGHQGKQ
QSEDEVTGGRQLTDDPREVARMLKRDNLASDIKIGDLPLIKNSEIQNFCLHGTVGAGKSEVIRRLMNYAR
KRGDMVIVYDRSCEFVKSYYDPAIDKILNPMDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLQNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDGRNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDIQTLPDLSCYVTLPGPYPAVKMAL
KYQERPKVAPEFIQRQIDTQAEQRLNAALAAREAESRSVAMLFEPDTEVVAPVSGPENAGQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQPQQPQPALVITPRPAPVAASTSAAAAAAVAGTGGTESVLETKAEEAEQL
PPGIDESGEVVDMAAWEAWQAEQDELTPQERQRREEVNINVHRTHDDDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIILYCLVILFGIIVALTLAVKLSWQTFVNGMAWYWCQTLAPMR
DIFRSQPVYTIRYYGKELKYNAQQLLQDKYVIWCADQVWTAFILAAIIAAVVCVIAFFVTTWILGHQGKQ
QSEDEVTGGRQLTDDPREVARMLKRDNLASDIKIGDLPLIKNSEIQNFCLHGTVGAGKSEVIRRLMNYAR
KRGDMVIVYDRSCEFVKSYYDPAIDKILNPMDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLQNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDGRNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDIQTLPDLSCYVTLPGPYPAVKMAL
KYQERPKVAPEFIQRQIDTQAEQRLNAALAAREAESRSVAMLFEPDTEVVAPVSGPENAGQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQPQQPQPALVITPRPAPVAASTSAAAAAAVAGTGGTESVLETKAEEAEQL
PPGIDESGEVVDMAAWEAWQAEQDELTPQERQRREEVNINVHRTHDDDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 58227..82000
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ENTER_RS22915 (ENTER_P0052) | 58227..60494 | - | 2268 | WP_202562370 | type IV conjugative transfer system coupling protein TraD | virb4 |
| ENTER_RS22920 (ENTER_P0054) | 60669..61400 | - | 732 | WP_202562372 | conjugal transfer complement resistance protein TraT | - |
| ENTER_RS22925 (ENTER_P0055) | 61491..61802 | + | 312 | WP_202562298 | heavy metal-binding domain-containing protein | - |
| ENTER_RS22930 (ENTER_P0056) | 61804..64662 | - | 2859 | WP_202562299 | conjugal transfer mating-pair stabilization protein TraG | traG |
| ENTER_RS22935 (ENTER_P0057) | 64662..66044 | - | 1383 | WP_202562300 | conjugal transfer pilus assembly protein TraH | traH |
| ENTER_RS22940 (ENTER_P0058) | 66031..66450 | - | 420 | WP_202562301 | conjugal transfer protein TrbF | - |
| ENTER_RS22945 (ENTER_P0059) | 66443..66991 | - | 549 | WP_202562302 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| ENTER_RS22950 (ENTER_P0060) | 66981..67226 | - | 246 | WP_102750541 | type-F conjugative transfer system pilin chaperone TraQ | - |
| ENTER_RS22955 (ENTER_P0061) | 67243..67950 | - | 708 | WP_202562381 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| ENTER_RS22960 (ENTER_P0062) | 68008..69804 | - | 1797 | WP_237658082 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| ENTER_RS22965 (ENTER_P0063) | 69864..70487 | - | 624 | WP_202562304 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| ENTER_RS22970 (ENTER_P0064) | 70502..71488 | - | 987 | WP_063449758 | conjugal transfer pilus assembly protein TraU | traU |
| ENTER_RS22975 (ENTER_P0065) | 71485..72132 | - | 648 | WP_202562305 | type-F conjugative transfer system protein TraW | traW |
| ENTER_RS22980 (ENTER_P0066) | 72132..72479 | - | 348 | WP_046622481 | type-F conjugative transfer system protein TrbI | - |
| ENTER_RS22985 (ENTER_P0067) | 72476..75106 | - | 2631 | WP_202562306 | type IV secretion system protein TraC | virb4 |
| ENTER_RS22990 (ENTER_P0069) | 75413..75949 | - | 537 | WP_202562307 | type IV conjugative transfer system lipoprotein TraV | traV |
| ENTER_RS22995 (ENTER_P0070) | 75970..77373 | - | 1404 | WP_202562308 | F-type conjugal transfer pilus assembly protein TraB | traB |
| ENTER_RS23000 (ENTER_P0071) | 77360..78103 | - | 744 | WP_202562309 | type-F conjugative transfer system secretin TraK | traK |
| ENTER_RS23005 (ENTER_P0072) | 78090..78656 | - | 567 | WP_202562310 | type IV conjugative transfer system protein TraE | traE |
| ENTER_RS23010 (ENTER_P0073) | 78671..78976 | - | 306 | WP_202562311 | type IV conjugative transfer system protein TraL | traL |
| ENTER_RS23015 (ENTER_P0074) | 79114..79464 | - | 351 | WP_202562312 | type IV conjugative transfer system pilin TraA | - |
| ENTER_RS23020 (ENTER_P0075) | 79511..79735 | - | 225 | WP_046622499 | TraY domain-containing protein | - |
| ENTER_RS23025 (ENTER_P0076) | 79814..80518 | - | 705 | WP_193037203 | LuxR C-terminal-related transcriptional regulator | - |
| ENTER_RS23030 (ENTER_P0077) | 80702..81097 | - | 396 | WP_202562313 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| ENTER_RS23035 (ENTER_P0079) | 81527..82000 | + | 474 | WP_202562314 | transglycosylase SLT domain-containing protein | virB1 |
| ENTER_RS23040 (ENTER_P0080) | 82041..82571 | - | 531 | WP_202562315 | antirestriction protein | - |
| ENTER_RS23045 (ENTER_P0081) | 83249..84079 | - | 831 | WP_202562316 | N-6 DNA methylase | - |
| ENTER_RS23050 (ENTER_P0082) | 84125..84388 | - | 264 | WP_046622508 | hypothetical protein | - |
| ENTER_RS23055 (ENTER_P0083) | 84474..84830 | - | 357 | WP_063449773 | hypothetical protein | - |
| ENTER_RS23060 (ENTER_P0084) | 84877..85158 | - | 282 | WP_237658073 | hypothetical protein | - |
| ENTER_RS23065 (ENTER_P0085) | 85220..85609 | - | 390 | WP_202562317 | ammonia monooxygenase | - |
| ENTER_RS23070 (ENTER_P0086) | 86310..86429 | - | 120 | WP_272929840 | type I toxin-antitoxin system Hok family toxin | - |
| ENTER_RS23460 (ENTER_P0087) | 86377..86583 | - | 207 | Protein_75 | DUF5431 family protein | - |
Host bacterium
| ID | 4256 | GenBank | NZ_LR881937 |
| Plasmid name | pUPC1 | Incompatibility group | IncFIB |
| Plasmid size | 121756 bp | Coordinate of oriT [Strand] | 81346..81458 [-] |
| Host baterium | Enterobacter cancerogenus strain UPC1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |