Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103811 |
| Name | oriT_pSymA |
| Organism | Sinorhizobium meliloti strain RMO17 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP009145 (998902..998930 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSymA
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file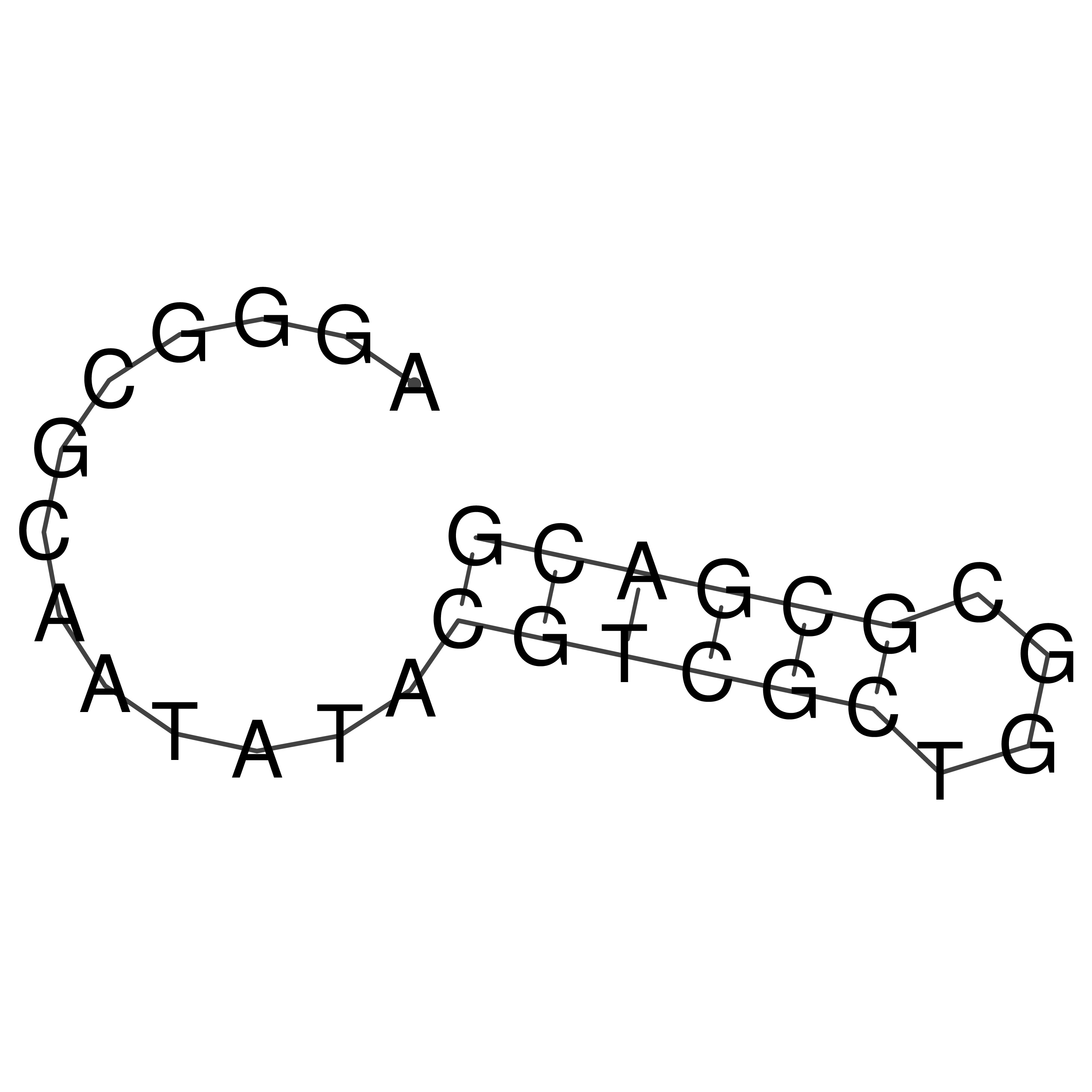
Relaxase
| ID | 2829 | GenBank | WP_040120788 |
| Name | traA_DU99_RS22245_pSymA |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 169780.54 Da Isoelectric Point: 9.4965
>WP_040120788.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAETGEAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRKQLNRADVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQHGRIVETETRAKIVDRIVADWADARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
ASLRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISQESYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDEQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFAALVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKRELVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVRKQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNMSERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAAERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRGITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSQEIISERQRRVIDRARSVTR
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAETGEAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRKQLNRADVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQHGRIVETETRAKIVDRIVADWADARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
ASLRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISQESYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDEQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFAALVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKRELVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVRKQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNMSERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAAERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRGITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSQEIISERQRRVIDRARSVTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2654 | GenBank | WP_014528597 |
| Name | traG_DU99_RS22260_pSymA |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70213.22 Da Isoelectric Point: 9.3518
>WP_014528597.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium meliloti]
MALKAKPHPSLLVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRKPVAMASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFDPNDPATWGQGGKVP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSIGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIEADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARSISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAVP
MALKAKPHPSLLVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRKPVAMASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFDPNDPATWGQGGKVP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSIGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIEADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARSISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 877111..886690
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| DU99_RS21595 (DU99_22425) | 873993..874841 | + | 849 | WP_040120779 | alpha/beta fold hydrolase | - |
| DU99_RS21600 (DU99_22430) | 874891..875538 | + | 648 | WP_010967700 | dihydrofolate reductase family protein | - |
| DU99_RS21605 (DU99_22435) | 875612..875863 | + | 252 | WP_010967699 | hypothetical protein | - |
| DU99_RS21610 (DU99_22440) | 876124..876492 | - | 369 | WP_010967698 | transcriptional regulator | - |
| DU99_RS36685 | 876589..877107 | + | 519 | Protein_872 | hypothetical protein | - |
| DU99_RS21615 (DU99_22445) | 877111..877782 | + | 672 | WP_010967696 | lytic transglycosylase domain-containing protein | virB1 |
| DU99_RS21620 (DU99_22450) | 877779..878078 | + | 300 | WP_010967695 | TrbC/VirB2 family protein | virB2 |
| DU99_RS21625 (DU99_22455) | 878083..878424 | + | 342 | WP_014528661 | type IV secretion system protein VirB3 | virB3 |
| DU99_RS21630 (DU99_22460) | 878399..880795 | + | 2397 | WP_014528660 | VirB4 family type IV secretion system protein | virb4 |
| DU99_RS21635 (DU99_22465) | 880795..881496 | + | 702 | WP_010967692 | P-type DNA transfer protein VirB5 | virB5 |
| DU99_RS21640 (DU99_22470) | 881493..881726 | + | 234 | WP_013845460 | EexN family lipoprotein | - |
| DU99_RS21645 (DU99_22475) | 881732..882667 | + | 936 | WP_014528659 | type IV secretion system protein | virB6 |
| DU99_RS21650 (DU99_22480) | 882664..882978 | + | 315 | WP_010967690 | hypothetical protein | - |
| DU99_RS21655 (DU99_22485) | 882983..883654 | + | 672 | WP_010967689 | virB8 family protein | virB8 |
| DU99_RS21660 (DU99_22490) | 883654..884508 | + | 855 | WP_014528658 | P-type conjugative transfer protein VirB9 | virB9 |
| DU99_RS21665 (DU99_22495) | 884518..885690 | + | 1173 | WP_010967687 | type IV secretion system protein VirB10 | virB10 |
| DU99_RS21670 (DU99_22500) | 885701..886690 | + | 990 | WP_010967686 | P-type DNA transfer ATPase VirB11 | virB11 |
| DU99_RS21675 (DU99_22505) | 887001..888227 | - | 1227 | WP_010967685 | MFS transporter | - |
| DU99_RS21680 (DU99_22510) | 888262..888873 | - | 612 | WP_010967684 | TetR/AcrR family transcriptional regulator | - |
| DU99_RS21685 (DU99_22515) | 889125..889801 | + | 677 | Protein_887 | YoaK family protein | - |
| DU99_RS21690 (DU99_22520) | 890049..891071 | + | 1023 | WP_010967682 | alcohol dehydrogenase AdhP | - |
Host bacterium
| ID | 4251 | GenBank | NZ_CP009145 |
| Plasmid name | pSymA | Incompatibility group | - |
| Plasmid size | 1466845 bp | Coordinate of oriT [Strand] | 998902..998930 [-] |
| Host baterium | Sinorhizobium meliloti strain RMO17 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | arsC, arsH, ncrA |
| Degradation gene | - |
| Symbiosis gene | fixA, fixB, fixN, fixO, fixG, fixS, nodM, nifN, nodD, nodA, nodB, nodC, nodI, nodJ, nodE, nodF, nodH, nifX, nifE, nifK, nifD, nifH, fixC, fixX, nifB, fixU |
| Anti-CRISPR | AcrIIA9 |