Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103802 |
| Name | oriT_pSTY1 |
| Organism | Salmonella enterica subsp. enterica serovar Typhimurium strain ATCC 13311 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP009103 (21190..21538 [+], 349 nt) |
| oriT length | 349 nt |
| IRs (inverted repeats) | 134..140, 154..160 (AAAATAT..ATATTTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 349 nt
>oriT_pSTY1
ACTGCGATGCAGGATAGGGCAAACGCCGCAAAATGACGTCTCTGACGCCATTCCGCAACGTTTGCCCGATGACCTACAGTCGGTTCTTGTTGGAGAGTTTTATGAAATATTTGGTCAAAGAGTTTATTAACGAAAAATATACTAAGGTTGTTAATATTTTAAAGGATAACCTTAAAGAACACTATCATGTTTTTTATTGTGTGAGATTAAGTGAGATTCTTTTTCCTGCCAGTGAATATGGAACTGAACAGTTTTTTAGTGATTTTGAAAAGATAAACTCCATTTCATTACCTGTCTTAGTGTTTGATCTCAAAGAGGGTGTTCCTGTGATTGTCATTAGTCTTGATGA
ACTGCGATGCAGGATAGGGCAAACGCCGCAAAATGACGTCTCTGACGCCATTCCGCAACGTTTGCCCGATGACCTACAGTCGGTTCTTGTTGGAGAGTTTTATGAAATATTTGGTCAAAGAGTTTATTAACGAAAAATATACTAAGGTTGTTAATATTTTAAAGGATAACCTTAAAGAACACTATCATGTTTTTTATTGTGTGAGATTAAGTGAGATTCTTTTTCCTGCCAGTGAATATGGAACTGAACAGTTTTTTAGTGATTTTGAAAAGATAAACTCCATTTCATTACCTGTCTTAGTGTTTGATCTCAAAGAGGGTGTTCCTGTGATTGTCATTAGTCTTGATGA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file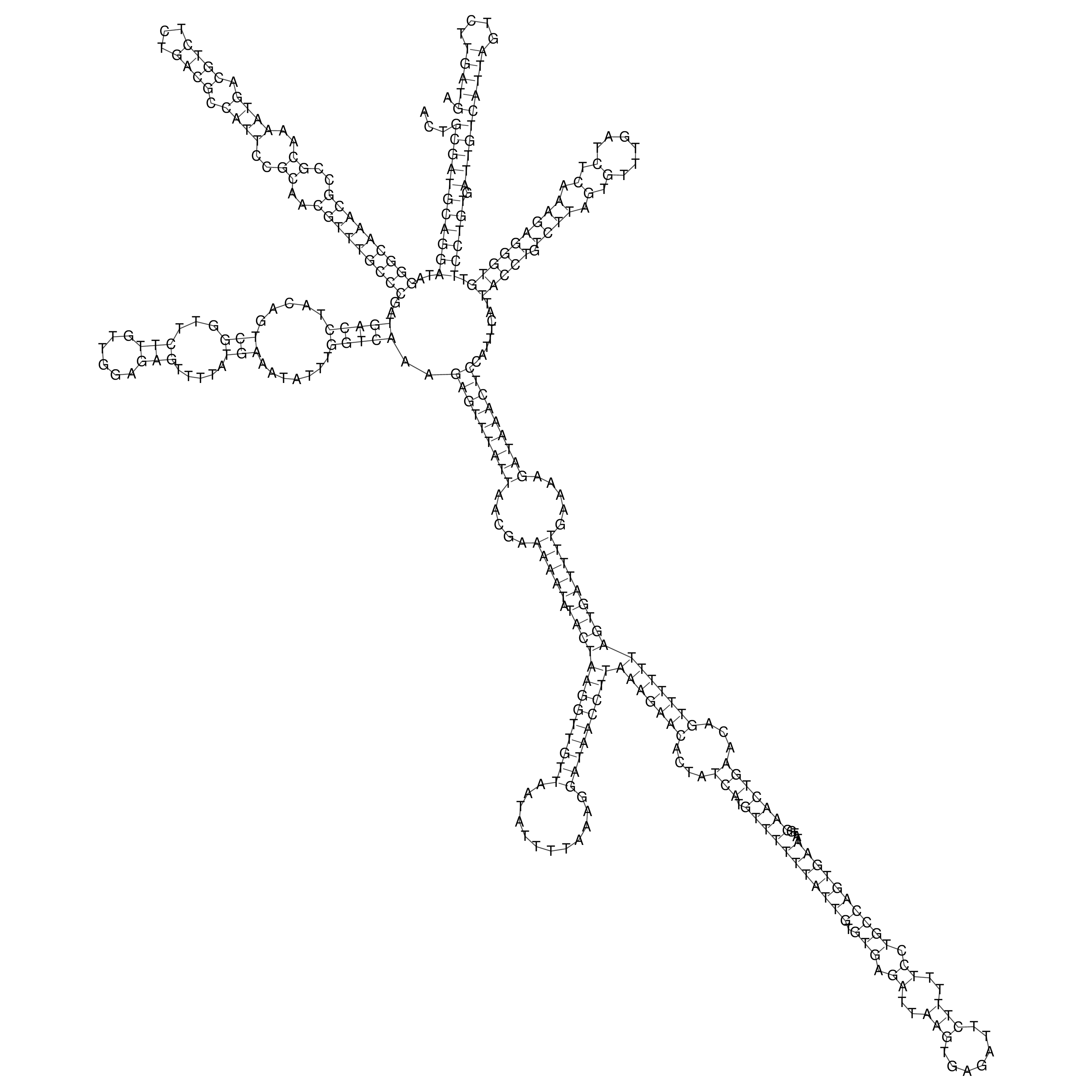
Relaxase
| ID | 2823 | GenBank | WP_025840297 |
| Name | Relaxase_IZ85_RS23165_pSTY1 |
UniProt ID | A0A6Y4C744 |
| Length | 388 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 388 a.a. Molecular weight: 44449.80 Da Isoelectric Point: 10.5368
>WP_025840297.1 MobP1 family relaxase [Salmonella enterica]
MGVYVDKEYRVKRKSSENGRKSAFAHKVKNGGKNYSRNVQERINRKGASKEVVVKISGGAVTRQGIRNSI
DYMSRESELPVMSESGRVWTGDEILEAKEHMIDRANDPQHVMNDKGKENKKITQNIVFSPPVSAKAKPED
LLESVRKTMQKKYPNHRFVLGYHCDKKEHPHVHVVFRIRDNDGKRADIRKKDLREIRTGFCEELKLKGYD
VKATHKQQHGLNQSVKDAHNTAPKRQKGVYEVVDIGYDHYQNDKTKSKQHFIKLKTLNKGVEKTYWGADF
GGLCSRESVKAGDLVRLKKLGQKEVKIPALDKNGVQHGWKTVHRNEWQLENLGVKGVDRTPSASKELVLN
SPDMLLKQQQRMAQFTQQKASTLQSEQKLKTGIKFWSL
MGVYVDKEYRVKRKSSENGRKSAFAHKVKNGGKNYSRNVQERINRKGASKEVVVKISGGAVTRQGIRNSI
DYMSRESELPVMSESGRVWTGDEILEAKEHMIDRANDPQHVMNDKGKENKKITQNIVFSPPVSAKAKPED
LLESVRKTMQKKYPNHRFVLGYHCDKKEHPHVHVVFRIRDNDGKRADIRKKDLREIRTGFCEELKLKGYD
VKATHKQQHGLNQSVKDAHNTAPKRQKGVYEVVDIGYDHYQNDKTKSKQHFIKLKTLNKGVEKTYWGADF
GGLCSRESVKAGDLVRLKKLGQKEVKIPALDKNGVQHGWKTVHRNEWQLENLGVKGVDRTPSASKELVLN
SPDMLLKQQQRMAQFTQQKASTLQSEQKLKTGIKFWSL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A6Y4C744 |
T4CP
| ID | 2646 | GenBank | WP_000053830 |
| Name | t4cp2_IZ85_RS23105_pSTY1 |
UniProt ID | _ |
| Length | 611 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 611 a.a. Molecular weight: 69403.32 Da Isoelectric Point: 7.1383
>WP_000053830.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacteriaceae]
MSLKLPDKGQWVFIGLVMCLVTYYAGSVAVYFLNGKTPLYIWKNFDSMLLWRIITESNIRTDIRLTAIPS
LLSGMVSSLIVPVFIIWQLNKTDVALYGDAKFACDNDLRKSKLLKWEKENDTDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREILLGNKVYLLDPFNSKTHQFNPLF
YIDLKAESGAKDLLKLIEILFPSYGMTGAEAHFNNLAGQYWTGLAKLLHFFINYEPSWLNEFGLKPVFSI
GSVVDLYSNIDRELILSKREELEGTNGLDENALYHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDITVYVGVNAEDISLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGIEGAKTLMSAHPCRIIYAVSEEDDAAKIS
EKLGYITTTSKSTSKSRGRSTSQGESESEARRALVLPQELGTLDFKEEFIILKGENPVKAEKALYYLDPY
FMDRLMKVSPKLASLTMELNKTKKIFGVKGLKYPSKEKMLSVGELESEVLL
MSLKLPDKGQWVFIGLVMCLVTYYAGSVAVYFLNGKTPLYIWKNFDSMLLWRIITESNIRTDIRLTAIPS
LLSGMVSSLIVPVFIIWQLNKTDVALYGDAKFACDNDLRKSKLLKWEKENDTDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREILLGNKVYLLDPFNSKTHQFNPLF
YIDLKAESGAKDLLKLIEILFPSYGMTGAEAHFNNLAGQYWTGLAKLLHFFINYEPSWLNEFGLKPVFSI
GSVVDLYSNIDRELILSKREELEGTNGLDENALYHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDITVYVGVNAEDISLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGIEGAKTLMSAHPCRIIYAVSEEDDAAKIS
EKLGYITTTSKSTSKSRGRSTSQGESESEARRALVLPQELGTLDFKEEFIILKGENPVKAEKALYYLDPY
FMDRLMKVSPKLASLTMELNKTKKIFGVKGLKYPSKEKMLSVGELESEVLL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3091..13044
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| IZ85_RS25675 | 237..431 | - | 195 | Protein_1 | ATP-binding protein | - |
| IZ85_RS24865 | 544..846 | - | 303 | WP_000820912 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| IZ85_RS24870 | 843..1256 | - | 414 | WP_000722606 | cag pathogenicity island Cag12 family protein | - |
| IZ85_RS23105 (IZ85_23070) | 1253..3088 | - | 1836 | WP_000053830 | type IV secretory system conjugative DNA transfer family protein | - |
| IZ85_RS23110 (IZ85_23075) | 3091..4122 | - | 1032 | WP_021557527 | P-type DNA transfer ATPase VirB11 | virB11 |
| IZ85_RS23115 (IZ85_23080) | 4124..5329 | - | 1206 | WP_000139139 | VirB10/TraB/TrbI family type IV secretion system protein | virB10 |
| IZ85_RS23120 (IZ85_23085) | 5326..6255 | - | 930 | WP_000783380 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| IZ85_RS23125 (IZ85_23090) | 6260..6973 | - | 714 | WP_000394613 | type IV secretion system protein | virB8 |
| IZ85_RS23130 (IZ85_23095) | 7219..8280 | - | 1062 | WP_040121201 | type IV secretion system protein | virB6 |
| IZ85_RS23135 (IZ85_23100) | 8292..8576 | - | 285 | WP_025840300 | EexN family lipoprotein | - |
| IZ85_RS23140 (IZ85_23105) | 8591..9340 | - | 750 | WP_025840299 | type IV secretion system protein | - |
| IZ85_RS23145 (IZ85_23110) | 9351..12104 | - | 2754 | WP_040121202 | VirB3 family type IV secretion system protein | virb4 |
| IZ85_RS23150 (IZ85_23115) | 12123..12419 | - | 297 | WP_025840298 | TrbC/VirB2 family protein | virB2 |
| IZ85_RS23155 (IZ85_23120) | 12397..13044 | - | 648 | WP_000953526 | lytic transglycosylase domain-containing protein | virB1 |
| IZ85_RS24875 | 13219..13737 | - | 519 | WP_001446885 | transcription termination/antitermination NusG family protein | - |
| IZ85_RS23165 (IZ85_23130) | 14090..15256 | - | 1167 | WP_025840297 | MobP1 family relaxase | - |
| IZ85_RS23170 (IZ85_23135) | 15259..15804 | - | 546 | WP_000757692 | DNA distortion polypeptide 1 | - |
| IZ85_RS24880 | 16130..16402 | - | 273 | WP_000160399 | hypothetical protein | - |
| IZ85_RS25685 | 16415..16564 | - | 150 | WP_000003880 | hypothetical protein | - |
| IZ85_RS24885 | 16634..16813 | - | 180 | WP_071881863 | hypothetical protein | - |
| IZ85_RS23180 (IZ85_23150) | 16856..17185 | - | 330 | WP_000866645 | hypothetical protein | - |
| IZ85_RS23185 (IZ85_23155) | 17278..17493 | - | 216 | WP_001180117 | hypothetical protein | - |
| IZ85_RS23190 (IZ85_23160) | 17483..17728 | - | 246 | WP_025840296 | hypothetical protein | - |
Host bacterium
| ID | 4242 | GenBank | NZ_CP009103 |
| Plasmid name | pSTY1 | Incompatibility group | IncX3 |
| Plasmid size | 38457 bp | Coordinate of oriT [Strand] | 21190..21538 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar Typhimurium strain ATCC 13311 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |