Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103800 |
| Name | oriT_SL42|unnamed2 |
| Organism | Rhizobium sp. SL42 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP063399 (115144..115187 [-], 44 nt) |
| oriT length | 44 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 44 nt
>oriT_SL42|unnamed2
GGATCCGAAGGGCGCAATTATACGTCGCTGATGCGACACTCTGC
GGATCCGAAGGGCGCAATTATACGTCGCTGATGCGACACTCTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file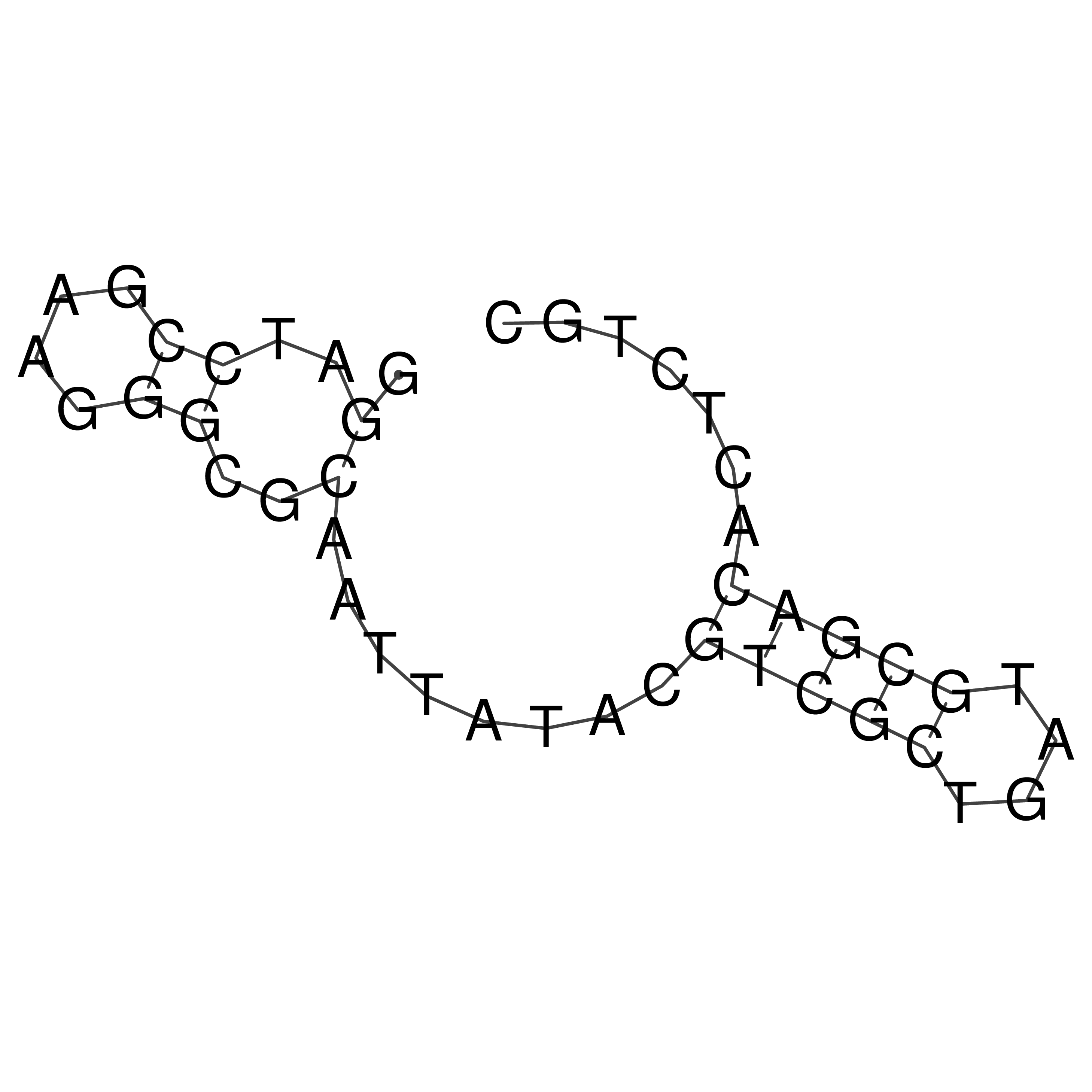
Relaxase
| ID | 2821 | GenBank | WP_237371934 |
| Name | traA_IM739_RS22840_SL42|unnamed2 |
UniProt ID | _ |
| Length | 1102 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1102 a.a. Molecular weight: 121733.48 Da Isoelectric Point: 9.5267
>WP_237371934.1 Ti-type conjugative transfer relaxase TraA [Rhizobium sp. SL42]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARAIDYRAKQGLLHEEFVIPSDAPEWLQSMIADR
SVSGASEAFWNKVEDFEKRSDAQLAKDVTIALPIELSPLQNIELVRDFVGRHITAEGMVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGSKKVAVTGPDGQPLRNDAGKIVYKLWAGGLENFNAFRDGWFACQNRHLAIAG
LDVRVDGRSFEEQGIELTPTIHLGVGAKAVERKADGKGEAASLERLELQEERRAENVRRIQRNPELVLDL
ITREKSVFDERDVAKVLHRYIDDAGLFQSLMARILQNPETLRLDRERLDFVTGVRAPEKYTTRELIRLEA
EMANRAILLSQRSSHGVRKPVLEATFARHERLSDEQKAAIEHVAGGERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELSWSQGRGMLDEKTVFVLDEAGMVSSRQMAMFVE
TVTRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDFARGHVGKAVAAYQ
SNGKMIGSELKADAVTNLIADWNREYDPARSTLILAHLRRDVRMLNELARAKLIERGIIEQGFVFKTADG
NRHFAAGDQIVFLKNEGAIGVKNGMLGKVVEAALGRIVAVIGEGEHLRQVIVEQRFYNNVDHGYATTVHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLGVYYGHRSFAKAGGLVEILSRKNSKETTLDYAGGSIY
SQALRFAEARGLHIVNVARTVARDRLEWTARQKQKLVDLAGRLAVLGARLGFATSAASTIQTSAKEAKPM
VAGITTFPKSIDQAVEDKLASDPALKKQWEEVSMRFHHVYAQPEAAFKAANLDAMLSDPATAAKTLSRIT
AEPEAFGSLKGKTGVFANRADKLDRDRALKNVRPLADSISDYLRQRGDTERRSHAEELAVRRQVALEIPA
LSSNARSVLERVRDAIDRNDLPSGLEFALADKMVKAELEGFAKAVTERFGERTFLPLAAKDANGEAFQRV
TSGMNAAQKSEVQQAWNTMRTVQQLSAHERSVTALKQTEALRQTRSQGLTLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARAIDYRAKQGLLHEEFVIPSDAPEWLQSMIADR
SVSGASEAFWNKVEDFEKRSDAQLAKDVTIALPIELSPLQNIELVRDFVGRHITAEGMVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGSKKVAVTGPDGQPLRNDAGKIVYKLWAGGLENFNAFRDGWFACQNRHLAIAG
LDVRVDGRSFEEQGIELTPTIHLGVGAKAVERKADGKGEAASLERLELQEERRAENVRRIQRNPELVLDL
ITREKSVFDERDVAKVLHRYIDDAGLFQSLMARILQNPETLRLDRERLDFVTGVRAPEKYTTRELIRLEA
EMANRAILLSQRSSHGVRKPVLEATFARHERLSDEQKAAIEHVAGGERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELSWSQGRGMLDEKTVFVLDEAGMVSSRQMAMFVE
TVTRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDFARGHVGKAVAAYQ
SNGKMIGSELKADAVTNLIADWNREYDPARSTLILAHLRRDVRMLNELARAKLIERGIIEQGFVFKTADG
NRHFAAGDQIVFLKNEGAIGVKNGMLGKVVEAALGRIVAVIGEGEHLRQVIVEQRFYNNVDHGYATTVHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLGVYYGHRSFAKAGGLVEILSRKNSKETTLDYAGGSIY
SQALRFAEARGLHIVNVARTVARDRLEWTARQKQKLVDLAGRLAVLGARLGFATSAASTIQTSAKEAKPM
VAGITTFPKSIDQAVEDKLASDPALKKQWEEVSMRFHHVYAQPEAAFKAANLDAMLSDPATAAKTLSRIT
AEPEAFGSLKGKTGVFANRADKLDRDRALKNVRPLADSISDYLRQRGDTERRSHAEELAVRRQVALEIPA
LSSNARSVLERVRDAIDRNDLPSGLEFALADKMVKAELEGFAKAVTERFGERTFLPLAAKDANGEAFQRV
TSGMNAAQKSEVQQAWNTMRTVQQLSAHERSVTALKQTEALRQTRSQGLTLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2644 | GenBank | WP_237371892 |
| Name | t4cp2_IM739_RS22585_SL42|unnamed2 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 92055.93 Da Isoelectric Point: 6.3776
>WP_237371892.1 conjugal transfer protein TrbE [Rhizobium sp. SL42]
MAALKRFRTTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDAESATDLERNELSRQINVILSRLG
TGWMIQVEAVRVPTVDYPSPDQCHFPDPVTRAIDAERRSHFQRAQGHFESKHAIILTYRPLESKKTAFAK
YIYSDEESRKKSYADTVLFVFRNAIREIEQYLANTLSIARMQTREVRERGGERTARYDELFQFVRFCITG
ESHPVRLPEVPMYLDWLVTGELQHGLTPKLENRFLAVVAIDGLPAESWPGILNNLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVVIL
FDKNREALQEKAEAIRRLLQAEGFGARVETLNATDAFLGSLPGNWYANIREPLINTSNLADLVPLNSVWS
GNPVNPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEFAQVFA
FDKGNSLLPLTLAAGGDHYEIGNDQSGEERALAFCPLFDLSTDGDRAWATGWLEMLVALQGVTITPDHRN
AISRQIGLMATAPGKSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLTLRAFQCFEIEQLMNMG
ERNLVPVLTYLFHRIEKRLDGSPSLILLDEAWLMLGHPVFRDKIREWLKVLRKTNCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNKRQIEIVSSAIPKREYYVATPDGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKTEHGQDWPAYWLESRGVHDATSHLNVR
MAALKRFRTTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDAESATDLERNELSRQINVILSRLG
TGWMIQVEAVRVPTVDYPSPDQCHFPDPVTRAIDAERRSHFQRAQGHFESKHAIILTYRPLESKKTAFAK
YIYSDEESRKKSYADTVLFVFRNAIREIEQYLANTLSIARMQTREVRERGGERTARYDELFQFVRFCITG
ESHPVRLPEVPMYLDWLVTGELQHGLTPKLENRFLAVVAIDGLPAESWPGILNNLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVVIL
FDKNREALQEKAEAIRRLLQAEGFGARVETLNATDAFLGSLPGNWYANIREPLINTSNLADLVPLNSVWS
GNPVNPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEFAQVFA
FDKGNSLLPLTLAAGGDHYEIGNDQSGEERALAFCPLFDLSTDGDRAWATGWLEMLVALQGVTITPDHRN
AISRQIGLMATAPGKSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLTLRAFQCFEIEQLMNMG
ERNLVPVLTYLFHRIEKRLDGSPSLILLDEAWLMLGHPVFRDKIREWLKVLRKTNCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNKRQIEIVSSAIPKREYYVATPDGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKTEHGQDWPAYWLESRGVHDATSHLNVR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2645 | GenBank | WP_237371937 |
| Name | traG_IM739_RS22855_SL42|unnamed2 |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 68832.65 Da Isoelectric Point: 8.2763
>WP_237371937.1 Ti-type conjugative transfer system protein TraG [Rhizobium sp. SL42]
MKANRIVLLILPVAMMIATVFVVTGIDTWLAGFGKTEAAKLGLGRAGVAAPYMAAAALGLIFLFAAAGSA
SIRAAGWGAATGNATIILTACAREGLRLAALSSQVPAGQSALSYLDPATMIGAAAALMSGCFAMRVAMVG
NAAFARAAPKRIVGKRALHGEADWMKLAEAERLFSQAGGIVIGERYRVDRDSVAGVSFRSENKETWGAGG
QSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLIVLDPSNEVAPMVSKHREDAGRRIRILD
PREPATGFNALDWVGQYGGTKEEDIASVASWIMSDSGRPTGVRDDFFRASALQLLTAIIADVCLSGHTPE
EQQTLRQVRVNLSEPEPKLRQRLQDIYDTSGSDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYQNYA
ALVSGSTFQTDDIASGTTDVFINIDLKTLETHAGLVRVVIGSFLNAIYNRDGAMEGRALFLLDEVARLGY
MRILETARDAGRKYSITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQSRGSSRTRSKQLAARPLIQPHEVLCMRADEQIIFTAGNPPLRCGRAIWFRREDMKSCVD
ANLFMKAGA
MKANRIVLLILPVAMMIATVFVVTGIDTWLAGFGKTEAAKLGLGRAGVAAPYMAAAALGLIFLFAAAGSA
SIRAAGWGAATGNATIILTACAREGLRLAALSSQVPAGQSALSYLDPATMIGAAAALMSGCFAMRVAMVG
NAAFARAAPKRIVGKRALHGEADWMKLAEAERLFSQAGGIVIGERYRVDRDSVAGVSFRSENKETWGAGG
QSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLIVLDPSNEVAPMVSKHREDAGRRIRILD
PREPATGFNALDWVGQYGGTKEEDIASVASWIMSDSGRPTGVRDDFFRASALQLLTAIIADVCLSGHTPE
EQQTLRQVRVNLSEPEPKLRQRLQDIYDTSGSDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYQNYA
ALVSGSTFQTDDIASGTTDVFINIDLKTLETHAGLVRVVIGSFLNAIYNRDGAMEGRALFLLDEVARLGY
MRILETARDAGRKYSITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQSRGSSRTRSKQLAARPLIQPHEVLCMRADEQIIFTAGNPPLRCGRAIWFRREDMKSCVD
ANLFMKAGA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 61254..70753
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| IM739_RS22550 (IM739_22535) | 56256..57470 | - | 1215 | WP_237371885 | plasmid replication protein RepC | - |
| IM739_RS22555 (IM739_22540) | 57627..58625 | - | 999 | WP_237371886 | plasmid partitioning protein RepB | - |
| IM739_RS22560 (IM739_22545) | 58622..59857 | - | 1236 | WP_237371887 | plasmid partitioning protein RepA | - |
| IM739_RS22565 (IM739_22550) | 60625..61251 | + | 627 | WP_237371888 | acyl-homoserine-lactone synthase | - |
| IM739_RS22570 (IM739_22555) | 61254..62225 | + | 972 | WP_237371889 | P-type conjugative transfer ATPase TrbB | virB11 |
| IM739_RS22575 (IM739_22560) | 62215..62589 | + | 375 | WP_237371890 | TrbC/VirB2 family protein | virB2 |
| IM739_RS22580 (IM739_22565) | 62589..62873 | + | 285 | WP_237371891 | conjugal transfer protein TrbD | virB3 |
| IM739_RS22585 (IM739_22570) | 62884..65340 | + | 2457 | WP_237371892 | conjugal transfer protein TrbE | virb4 |
| IM739_RS22590 (IM739_22575) | 65312..66115 | + | 804 | WP_237371893 | P-type conjugative transfer protein TrbJ | virB5 |
| IM739_RS22595 (IM739_22580) | 66112..66315 | + | 204 | WP_237371894 | entry exclusion protein TrbK | - |
| IM739_RS22600 (IM739_22585) | 66309..67487 | + | 1179 | WP_237371895 | P-type conjugative transfer protein TrbL | virB6 |
| IM739_RS22605 (IM739_22590) | 67509..68171 | + | 663 | WP_237371896 | conjugal transfer protein TrbF | virB8 |
| IM739_RS22610 (IM739_22595) | 68188..69000 | + | 813 | WP_237371897 | P-type conjugative transfer protein TrbG | virB9 |
| IM739_RS22615 (IM739_22600) | 69004..69441 | + | 438 | WP_237371898 | conjugal transfer protein TrbH | - |
| IM739_RS22620 (IM739_22605) | 69455..70753 | + | 1299 | WP_237371899 | IncP-type conjugal transfer protein TrbI | virB10 |
| IM739_RS22625 (IM739_22610) | 70991..72232 | + | 1242 | WP_237371900 | glycosyltransferase | - |
| IM739_RS22630 (IM739_22615) | 72413..72886 | - | 474 | WP_237371901 | isoprenylcysteine carboxylmethyltransferase family protein | - |
| IM739_RS22635 (IM739_22620) | 72923..73302 | - | 380 | Protein_63 | antibiotic biosynthesis monooxygenase family protein | - |
| IM739_RS22640 (IM739_22625) | 73358..74251 | - | 894 | WP_237371902 | NAD(P)/FAD-dependent oxidoreductase | - |
| IM739_RS22645 (IM739_22630) | 74469..75116 | + | 648 | WP_237371903 | TetR/AcrR family transcriptional regulator | - |
Host bacterium
| ID | 4240 | GenBank | NZ_CP063399 |
| Plasmid name | SL42|unnamed2 | Incompatibility group | - |
| Plasmid size | 351829 bp | Coordinate of oriT [Strand] | 115144..115187 [-] |
| Host baterium | Rhizobium sp. SL42 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixA, fixB |
| Anti-CRISPR | - |