Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103790 |
| Name | oriT_pTB44P3 |
| Organism | Escherichia fergusonii strain EF44 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP070957 (22689..22741 [+], 53 nt) |
| oriT length | 53 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 53 nt
>oriT_pTB44P3
CACACGATTGTAACATGACCGGAACGGTCTTGTGTACAATCGGTATCGTGCCT
CACACGATTGTAACATGACCGGAACGGTCTTGTGTACAATCGGTATCGTGCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file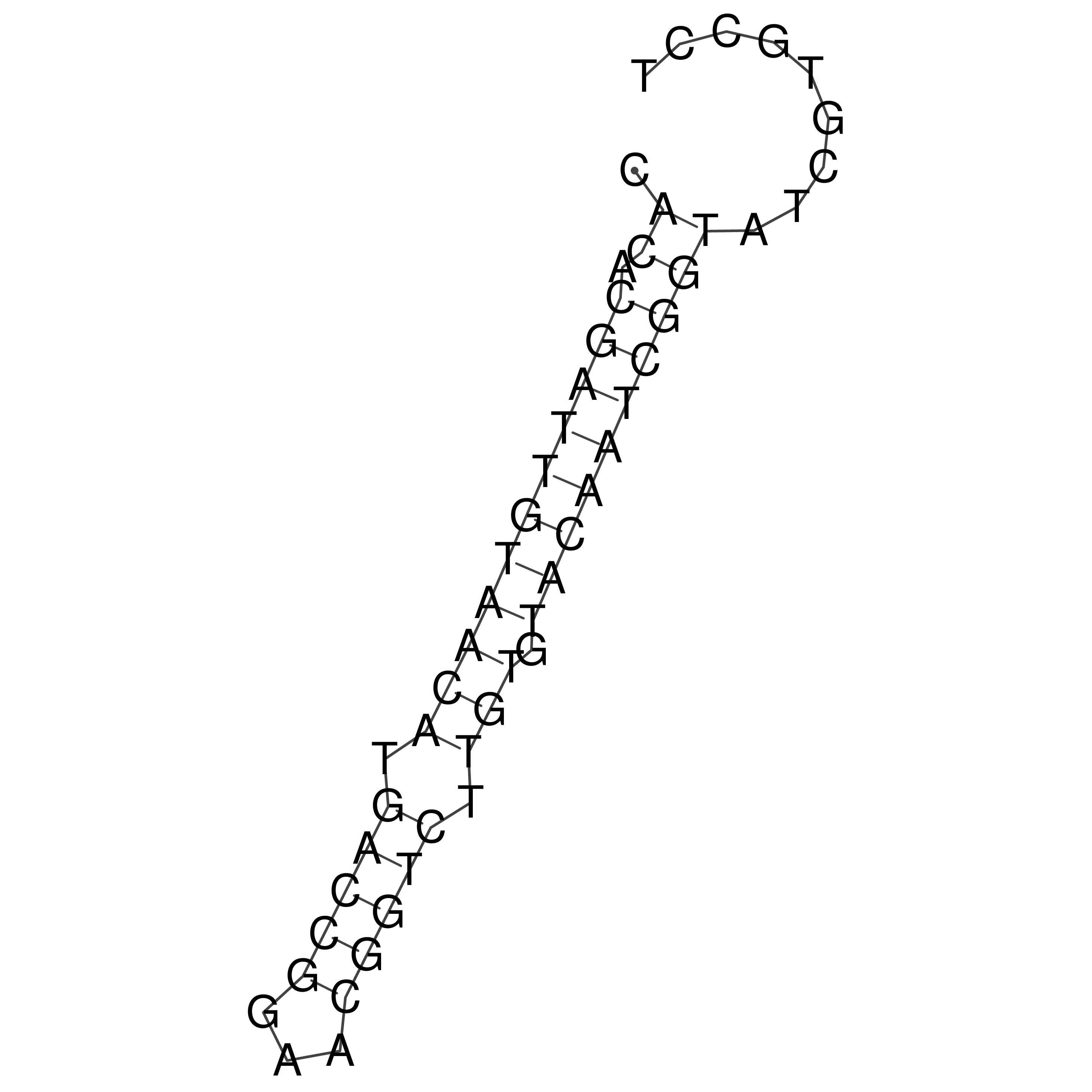
T4CP
| ID | 2639 | GenBank | WP_015059539 |
| Name | t4cp2_JYG65_RS22995_pTB44P3 |
UniProt ID | _ |
| Length | 652 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 652 a.a. Molecular weight: 73404.02 Da Isoelectric Point: 9.4339
>WP_015059539.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacteriaceae]
MNAKKMGGLILFLLLLLVGVLIASNYLGGYTALRYSSVDMSLLKWDTFHSVISTFSGNPQYKKLVFMAWF
GFSVPLIFFAIFMLIVVIGIMPKKVIYGDARLATDMDLSKSGFFPDKKSPYKHPPILIGKMFKGRYKKQF
IYFAGQQFLILYAPTRSGKGVGIVIPNCVNYPGSMVILDIKLENWFLSAGFRQKELGQKCFLFAPAGYAE
TIDQAIKGQIRSHRWNPLDCVSRSDLLRETDLAKIAAILIPASDDPIWSDSARNLFVGLGLYLLDKERFH
LDQKAKGHNAPDVLVSISAILKTSIPDNGKDLAAWMGQEVENRSWISDKTKSFFFEFMSAPDRTRGSIKT
NFSSPLNIFSNPVTAEATNFSDFDIRDIRKKPMSIYLGLTPDALITHEKIVNLFFSLLVNENCRELPEHN
PDLKYQCLILLDEFTSMGKSEVIERAVGFTAGYNLRFMFILQNEGQGQKSDMYGQEGWTTFTENSAVVLY
YPPKSKNALAKKISEEIGVRDMKISKRSISSGGGKGGSSRTRNDDVIERPVLLPEEIVSLRDKKNKARNI
AIREIITSEFSRPFIANKIIWFEEPEFKRRVDIARNNHVDIPNLFTQEVMDEIAKIAEIYLPKAGGKKVM
VAGGNVITNPDLDNHDKTDVSE
MNAKKMGGLILFLLLLLVGVLIASNYLGGYTALRYSSVDMSLLKWDTFHSVISTFSGNPQYKKLVFMAWF
GFSVPLIFFAIFMLIVVIGIMPKKVIYGDARLATDMDLSKSGFFPDKKSPYKHPPILIGKMFKGRYKKQF
IYFAGQQFLILYAPTRSGKGVGIVIPNCVNYPGSMVILDIKLENWFLSAGFRQKELGQKCFLFAPAGYAE
TIDQAIKGQIRSHRWNPLDCVSRSDLLRETDLAKIAAILIPASDDPIWSDSARNLFVGLGLYLLDKERFH
LDQKAKGHNAPDVLVSISAILKTSIPDNGKDLAAWMGQEVENRSWISDKTKSFFFEFMSAPDRTRGSIKT
NFSSPLNIFSNPVTAEATNFSDFDIRDIRKKPMSIYLGLTPDALITHEKIVNLFFSLLVNENCRELPEHN
PDLKYQCLILLDEFTSMGKSEVIERAVGFTAGYNLRFMFILQNEGQGQKSDMYGQEGWTTFTENSAVVLY
YPPKSKNALAKKISEEIGVRDMKISKRSISSGGGKGGSSRTRNDDVIERPVLLPEEIVSLRDKKNKARNI
AIREIITSEFSRPFIANKIIWFEEPEFKRRVDIARNNHVDIPNLFTQEVMDEIAKIAEIYLPKAGGKKVM
VAGGNVITNPDLDNHDKTDVSE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 42441..58610
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| JYG65_RS22850 (JYG65_22755) | 38353..38805 | + | 453 | WP_000101552 | CaiF/GrlA family transcriptional regulator | - |
| JYG65_RS22855 (JYG65_22760) | 38798..39013 | + | 216 | WP_001127357 | DUF1187 family protein | - |
| JYG65_RS22860 (JYG65_22765) | 39006..39182 | + | 177 | WP_000753050 | hypothetical protein | - |
| JYG65_RS22865 (JYG65_22770) | 39209..40162 | + | 954 | WP_072089442 | SPFH domain-containing protein | - |
| JYG65_RS22870 (JYG65_22775) | 40536..40979 | + | 444 | WP_000964330 | NfeD family protein | - |
| JYG65_RS22875 (JYG65_22780) | 40983..41153 | + | 171 | WP_000550720 | hypothetical protein | - |
| JYG65_RS22880 (JYG65_22785) | 41164..41610 | + | 447 | WP_001243165 | hypothetical protein | - |
| JYG65_RS22885 (JYG65_22790) | 41643..41900 | + | 258 | WP_001542015 | hypothetical protein | - |
| JYG65_RS22890 (JYG65_22795) | 41881..42183 | + | 303 | WP_001360345 | hypothetical protein | - |
| JYG65_RS22895 (JYG65_22800) | 42180..42437 | + | 258 | WP_000739144 | hypothetical protein | - |
| JYG65_RS22900 (JYG65_22805) | 42441..43436 | + | 996 | WP_001028543 | type IV secretion system protein | virB6 |
| JYG65_RS22905 (JYG65_22810) | 43442..44083 | + | 642 | WP_001425343 | type IV secretion system protein | - |
| JYG65_RS22910 (JYG65_22815) | 44085..44339 | + | 255 | WP_001043555 | EexN family lipoprotein | - |
| JYG65_RS22915 (JYG65_22820) | 44352..44639 | + | 288 | WP_001032611 | EexN family lipoprotein | - |
| JYG65_RS22920 (JYG65_22825) | 44712..45347 | + | 636 | WP_015059536 | hypothetical protein | - |
| JYG65_RS22925 (JYG65_22830) | 45447..45698 | + | 252 | WP_000121741 | hypothetical protein | - |
| JYG65_RS22930 (JYG65_22835) | 45688..45969 | + | 282 | WP_000638823 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| JYG65_RS22935 (JYG65_22840) | 46037..46336 | + | 300 | WP_000835764 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| JYG65_RS22940 (JYG65_22845) | 46339..47574 | + | 1236 | WP_015059538 | TcpQ domain-containing protein | - |
| JYG65_RS22945 (JYG65_22850) | 47580..48017 | + | 438 | WP_000539665 | type IV pilus biogenesis protein PilM | - |
| JYG65_RS22950 (JYG65_22855) | 48136..48534 | + | 399 | WP_001153665 | hypothetical protein | - |
| JYG65_RS22955 (JYG65_22860) | 48555..49139 | + | 585 | WP_001177113 | lytic transglycosylase domain-containing protein | virB1 |
| JYG65_RS22960 | 49139..49429 | + | 291 | WP_000865479 | conjugal transfer protein | - |
| JYG65_RS22965 (JYG65_22870) | 49500..49820 | + | 321 | WP_000362080 | VirB3 family type IV secretion system protein | virB3 |
| JYG65_RS22970 (JYG65_22875) | 49826..52183 | + | 2358 | WP_063078682 | VirB4 family type IV secretion system protein | virb4 |
| JYG65_RS22975 (JYG65_22880) | 52349..53083 | + | 735 | WP_000432282 | type IV secretion system protein | virB8 |
| JYG65_RS22980 (JYG65_22885) | 53149..53850 | + | 702 | WP_000274524 | TrbG/VirB9 family P-type conjugative transfer protein | - |
| JYG65_RS22985 (JYG65_22890) | 53840..54979 | + | 1140 | WP_000790640 | TrbI/VirB10 family protein | virB10 |
| JYG65_RS22990 (JYG65_22895) | 54998..56053 | + | 1056 | WP_001059977 | P-type DNA transfer ATPase VirB11 | virB11 |
| JYG65_RS22995 (JYG65_22900) | 56069..58027 | + | 1959 | WP_015059539 | type IV secretory system conjugative DNA transfer family protein | - |
| JYG65_RS23000 (JYG65_22905) | 58074..58610 | + | 537 | WP_001220543 | sigma 54-interacting transcriptional regulator | virb4 |
| JYG65_RS23005 (JYG65_22910) | 58603..60246 | + | 1644 | WP_001035592 | PilN family type IVB pilus formation outer membrane protein | - |
| JYG65_RS23010 (JYG65_22915) | 60297..60713 | + | 417 | WP_223674109 | type 4b pilus protein PilO2 | - |
| JYG65_RS23015 (JYG65_22920) | 60739..61952 | + | 1214 | WP_162829202 | IS3-like element IS1203 family transposase | - |
Host bacterium
| ID | 4230 | GenBank | NZ_CP070957 |
| Plasmid name | pTB44P3 | Incompatibility group | IncI2 |
| Plasmid size | 62882 bp | Coordinate of oriT [Strand] | 22689..22741 [+] |
| Host baterium | Escherichia fergusonii strain EF44 |
Cargo genes
| Drug resistance gene | mcr-1.1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |