Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103759 |
| Name | oriT_p712 |
| Organism | Ralstonia pickettii |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_019318 (32879..32953 [-], 75 nt) |
| oriT length | 75 nt |
| IRs (inverted repeats) | 25..31, 37..43 (TAACCGG..CCGGTTA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 75 nt
>oriT_p712
CGAATAAGGGACAGTGAAGATAGATAACCGGCTTGCCCGGTTAGCTAACTTCACCCATCCTGCCCGCCTTACGGC
CGAATAAGGGACAGTGAAGATAGATAACCGGCTTGCCCGGTTAGCTAACTTCACCCATCCTGCCCGCCTTACGGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file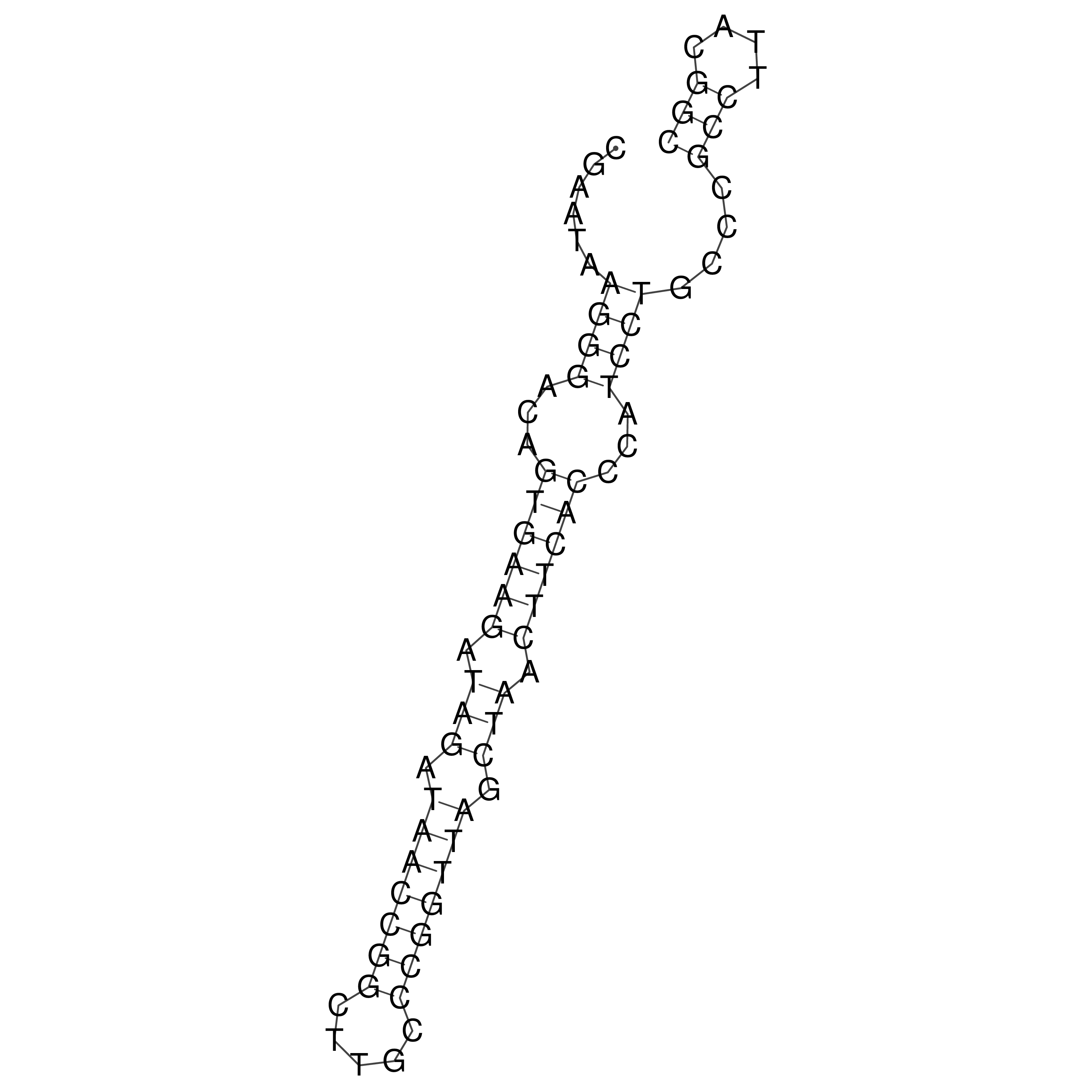
Relaxase
| ID | 2791 | GenBank | WP_015061637 |
| Name | Relaxase_HS994_RS00190_p712 |
UniProt ID | I3RYN8 |
| Length | 740 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 740 a.a. Molecular weight: 81671.57 Da Isoelectric Point: 10.6618
>WP_015061637.1 TraI/MobA(P) family conjugative relaxase [Ralstonia pickettii]
MIAKHVPMRSLGKSDFAGLAEYITDAQSKTERLGLVTVTNCQAGTVQAATDEVLATQHMNTRATGDKTFH
LLVSFRAGEKPDTDTLKAIEERICAGLGYGEHQRISAVHHDTDNLHIHIAINKIHPTRNTMHEPFQSYRT
LGELCEVLERDYGLEKDNHTPARSIAEGRAADMERHAGVESLVGWIKRECLEEIKAAQSWTELHEALQAN
GLELRAKGNGFVIEAGDGTQVKASTVARELSKPKLEARFGPFEASPERQAETKAKRQYQKKPVRLRVNTV
ELYARYKADQQTLAATQRAALDKARQRKTRKVEDAKRSNRLRRATIKVLGENRLNKKLLYAQASKALRAE
IDSINKQYQKDRQDCYDAHKRRTWADWLKKEALQGNSEALAALRAREAAQGLKGNTVKGEGQAKPGHAPV
VDNITKKGTIIFRAGKTAVRDDGDKLQVSREATREGLQQALRLAMERYGNRITVNGTAAFKAQIIRAAVD
SQLPITFADPALESRRQALLKKENTHDRPEQDRGRTGRGTGRAGSRPAADDNATRSGINAGGSGRNPAGG
IHAAGQHRKPDIGRIGRVPPPQRKNRLRALSKLGVVRIASGSEVLLPRDVPGHMEQQGTQPDNALRRGVS
GPGGRLDARPEYKPEQVKAMDKYIAEREGKRLKGFDIPKHSRYTDGYGALSFAGMRNVEGQDLALVRRGD
DVMVMPIDQATARRLKRIAIGDAVSITPKGSIKTSKGRSR
MIAKHVPMRSLGKSDFAGLAEYITDAQSKTERLGLVTVTNCQAGTVQAATDEVLATQHMNTRATGDKTFH
LLVSFRAGEKPDTDTLKAIEERICAGLGYGEHQRISAVHHDTDNLHIHIAINKIHPTRNTMHEPFQSYRT
LGELCEVLERDYGLEKDNHTPARSIAEGRAADMERHAGVESLVGWIKRECLEEIKAAQSWTELHEALQAN
GLELRAKGNGFVIEAGDGTQVKASTVARELSKPKLEARFGPFEASPERQAETKAKRQYQKKPVRLRVNTV
ELYARYKADQQTLAATQRAALDKARQRKTRKVEDAKRSNRLRRATIKVLGENRLNKKLLYAQASKALRAE
IDSINKQYQKDRQDCYDAHKRRTWADWLKKEALQGNSEALAALRAREAAQGLKGNTVKGEGQAKPGHAPV
VDNITKKGTIIFRAGKTAVRDDGDKLQVSREATREGLQQALRLAMERYGNRITVNGTAAFKAQIIRAAVD
SQLPITFADPALESRRQALLKKENTHDRPEQDRGRTGRGTGRAGSRPAADDNATRSGINAGGSGRNPAGG
IHAAGQHRKPDIGRIGRVPPPQRKNRLRALSKLGVVRIASGSEVLLPRDVPGHMEQQGTQPDNALRRGVS
GPGGRLDARPEYKPEQVKAMDKYIAEREGKRLKGFDIPKHSRYTDGYGALSFAGMRNVEGQDLALVRRGD
DVMVMPIDQATARRLKRIAIGDAVSITPKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | I3RYN8 |
T4CP
| ID | 2618 | GenBank | WP_031942766 |
| Name | t4cp2_HS994_RS00195_p712 |
UniProt ID | _ |
| Length | 634 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 634 a.a. Molecular weight: 69698.95 Da Isoelectric Point: 8.4143
>WP_031942766.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Pseudomonadota]
MKIKMNNAVGPQVRSAKPKPSKLLPTLGVLSLAGGLQASTQFFAHTFQYQASLGPNIGHVYAPWSILQWA
SKWYGHYPNEIMSAGSMGMLVTTVGLLGVAIAKVVASNTSKANEYLHGSARWAEKKDIQAAGLLPRPRTV
LEVVTGKEEATATGVYVGGWEDKDGSFYYLRHSGPEHVLCYAPTRSGKGVGLVVPTMLSWPASAVITDLK
GELWALTAGWRKKHAKNRVLRFEPASSSGGICWNPLDEIRLGTEFEVGDVQNLAQLIVDPDGKGLDSHWQ
KTAFSLLVGVILHALYKAKADGTYATLPSVDAMLADPNRDIGELWMEMTTYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLSLYRDPVVARSVSKSEFRIKDLMHQDDPASLYIVTQPNDKARLRPLVRIMVN
MIVRLLADKMDFEDGRPVAHYKHRLLMMLDEFPSLGKLEILQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDETITSNCHVQNAYPPNRVETAEHLSRLTGQTTIVKEQITTSGRRTAAMLGQVSRTFQEVQRPLLT
PDECLRMPGPRKSATGEIEQAGDMVVYVAGYPAIYGKQPLYFKDPVFQARAAIPAPKVSDKTIHIQDAGQ
GITI
MKIKMNNAVGPQVRSAKPKPSKLLPTLGVLSLAGGLQASTQFFAHTFQYQASLGPNIGHVYAPWSILQWA
SKWYGHYPNEIMSAGSMGMLVTTVGLLGVAIAKVVASNTSKANEYLHGSARWAEKKDIQAAGLLPRPRTV
LEVVTGKEEATATGVYVGGWEDKDGSFYYLRHSGPEHVLCYAPTRSGKGVGLVVPTMLSWPASAVITDLK
GELWALTAGWRKKHAKNRVLRFEPASSSGGICWNPLDEIRLGTEFEVGDVQNLAQLIVDPDGKGLDSHWQ
KTAFSLLVGVILHALYKAKADGTYATLPSVDAMLADPNRDIGELWMEMTTYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLSLYRDPVVARSVSKSEFRIKDLMHQDDPASLYIVTQPNDKARLRPLVRIMVN
MIVRLLADKMDFEDGRPVAHYKHRLLMMLDEFPSLGKLEILQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDETITSNCHVQNAYPPNRVETAEHLSRLTGQTTIVKEQITTSGRRTAAMLGQVSRTFQEVQRPLLT
PDECLRMPGPRKSATGEIEQAGDMVVYVAGYPAIYGKQPLYFKDPVFQARAAIPAPKVSDKTIHIQDAGQ
GITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2619 | GenBank | WP_015061655 |
| Name | t4cp2_HS994_RS00300_p712 |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 94240.21 Da Isoelectric Point: 6.2436
>WP_015061655.1 MULTISPECIES: VirB4 family type IV secretion/conjugal transfer ATPase [Pseudomonadota]
MIESIAFAIAVLGAVLLLILFARIRAVDAELKLKKHRSKDAGLADLLNYAAMVDDGVIVGKNGSFMAAWL
YKGDDNASSTDEQREMVSFRINQALAGLGSGWMVHVDAVRRPAPNYSDRGHSTFPDPLSAAMDEERRQLF
QGLGTMYEGYFVLTVTWFPPLLAQRKFVELMFDDDAISPDHKARTMGLIKQFKRETTSIESRLSSAVKLT
RLRGQKVVTEEGREVTHDDFLSWLQFCITGLHHPVQLPSNPMYLDALIGGQEMFGGVVPKVGRNFVQVVA
VEGFPLESTPGVLTALGELACEYRWSSRFIFMDQHEAVAHLDKFRKKWRQKVRGFFDQVFNTNSGNIDQD
ALSMVGDAEAAIAEVNSGLVAVGYYTGVVVLMDEDRTRLEQSARQVEKAINRMGFAARIETINTLDAFLG
SLPGHGVENVRRPLINTLNLSDLLPTSTIWTGLANAPCPMYPPLAPALMHCVTHGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGLIAAQLRRYAGMSIFAFDKGMSMYPLAAGIRAASKGKSGLHFTVAADDDQLAFC
PLQFLETKGDRAWAMEWIDTILTLNGVETTPGQRNEIGNAILSMHASGAHTLSEFSVTIQDETIREAIRQ
YTVDGTMGHLLDAETDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALHGQPAVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVATIKSLEAKFGYAWVDEWLAG
RGLNLNDYLEAA
MIESIAFAIAVLGAVLLLILFARIRAVDAELKLKKHRSKDAGLADLLNYAAMVDDGVIVGKNGSFMAAWL
YKGDDNASSTDEQREMVSFRINQALAGLGSGWMVHVDAVRRPAPNYSDRGHSTFPDPLSAAMDEERRQLF
QGLGTMYEGYFVLTVTWFPPLLAQRKFVELMFDDDAISPDHKARTMGLIKQFKRETTSIESRLSSAVKLT
RLRGQKVVTEEGREVTHDDFLSWLQFCITGLHHPVQLPSNPMYLDALIGGQEMFGGVVPKVGRNFVQVVA
VEGFPLESTPGVLTALGELACEYRWSSRFIFMDQHEAVAHLDKFRKKWRQKVRGFFDQVFNTNSGNIDQD
ALSMVGDAEAAIAEVNSGLVAVGYYTGVVVLMDEDRTRLEQSARQVEKAINRMGFAARIETINTLDAFLG
SLPGHGVENVRRPLINTLNLSDLLPTSTIWTGLANAPCPMYPPLAPALMHCVTHGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGLIAAQLRRYAGMSIFAFDKGMSMYPLAAGIRAASKGKSGLHFTVAADDDQLAFC
PLQFLETKGDRAWAMEWIDTILTLNGVETTPGQRNEIGNAILSMHASGAHTLSEFSVTIQDETIREAIRQ
YTVDGTMGHLLDAETDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALHGQPAVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVATIKSLEAKFGYAWVDEWLAG
RGLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 49658..60283
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HS994_RS00220 | 44765..45097 | - | 333 | WP_172684580 | SMR family transporter | - |
| HS994_RS00225 | 45104..45643 | - | 540 | WP_172684581 | TetR/AcrR family transcriptional regulator | - |
| HS994_RS00230 (p712_0450) | 45853..46503 | + | 651 | WP_015061642 | recombinase family protein | - |
| HS994_RS00235 (p712_0460) | 46533..46940 | - | 408 | WP_015061643 | hypothetical protein | - |
| HS994_RS00240 (p712_0470) | 46985..47416 | - | 432 | WP_031942760 | OmpA family protein | - |
| HS994_RS00245 (p712_0480) | 47431..48129 | - | 699 | WP_015061645 | TraX family protein | - |
| HS994_RS00250 (p712_0490) | 48143..48400 | - | 258 | WP_031942758 | hypothetical protein | - |
| HS994_RS00255 (p712_0500) | 48397..49038 | - | 642 | WP_102636876 | transglycosylase SLT domain-containing protein | - |
| HS994_RS00260 (p712_0510) | 49053..49640 | - | 588 | WP_015061647 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| HS994_RS00265 (p712_0520) | 49658..51433 | - | 1776 | WP_031942757 | P-type conjugative transfer protein TrbL | virB6 |
| HS994_RS00270 (p712_0530) | 51441..51665 | - | 225 | WP_015061649 | entry exclusion lipoprotein TrbK | - |
| HS994_RS00275 (p712_0540) | 51675..52442 | - | 768 | WP_015061650 | P-type conjugative transfer protein TrbJ | virB5 |
| HS994_RS00280 (p712_0550) | 52460..53848 | - | 1389 | WP_015061651 | TrbI/VirB10 family protein | virB10 |
| HS994_RS00285 (p712_0560) | 53852..54301 | - | 450 | WP_015061652 | TrbH protein | - |
| HS994_RS00290 (p712_0570) | 54303..55196 | - | 894 | WP_015061653 | P-type conjugative transfer protein TrbG | virB9 |
| HS994_RS00295 (p712_0580) | 55215..55979 | - | 765 | WP_015061654 | conjugal transfer protein TrbF | virB8 |
| HS994_RS00300 (p712_0590) | 55976..58534 | - | 2559 | WP_015061655 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| HS994_RS00305 (p712_0600) | 58531..58842 | - | 312 | WP_015061656 | conjugal transfer protein TrbD | virB3 |
| HS994_RS00310 (p712_0610) | 58846..59307 | - | 462 | WP_015061657 | conjugal transfer system pilin TrbC | virB2 |
| HS994_RS00315 (p712_0620) | 59321..60283 | - | 963 | WP_015061658 | P-type conjugative transfer ATPase TrbB | virB11 |
| HS994_RS00320 (p712_0630) | 60569..60946 | - | 378 | WP_015061659 | transcriptional regulator | - |
| HS994_RS00325 (p712_0640) | 61063..61404 | + | 342 | WP_031942755 | single-stranded DNA-binding protein | - |
| HS994_RS00330 (p712_0650) | 61456..62691 | + | 1236 | WP_015061661 | plasmid replication initiator TrfA | - |
Host bacterium
| ID | 4199 | GenBank | NC_019318 |
| Plasmid name | p712 | Incompatibility group | - |
| Plasmid size | 62798 bp | Coordinate of oriT [Strand] | 32879..32953 [-] |
| Host baterium | Ralstonia pickettii |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |