Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103714 |
| Name | oriT_pSCD1 |
| Organism | Salmonella enterica subsp. enterica serovar Bovismorbificans strain SB699 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP091299 (39026..39295 [+], 270 nt) |
| oriT length | 270 nt |
| IRs (inverted repeats) | 245..253, 262..270 (CACCCCTGG..CCAGGGGTG) 92..98, 111..117 (ATTTAAT..ATTAAAT) 103..108, 111..116 (TTTAAT..ATTAAA) 31..38, 41..48 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 270 nt
GTCCTCACCACCAAAAGCACCACACCCCACGCAAAATTAAAATTTTGCTGATTTGTTTTTAGTAACAGTGAGTTATGTTCATAATAATGTAATTTAATTTATTTTAATGTATTAAATAGCGGTGCAGGCACGTCGCCGGCACCGCTATGACGACGCTATACAGGGGTAGTACTGACTATTTTTATAAAAAACATTGCATTATATCAGGGGCGCTGCTAGCGTCGCTGTGCCTGTTTTTAAGTAACACCCCTGGGGGCGCTGCCAGGGGTG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file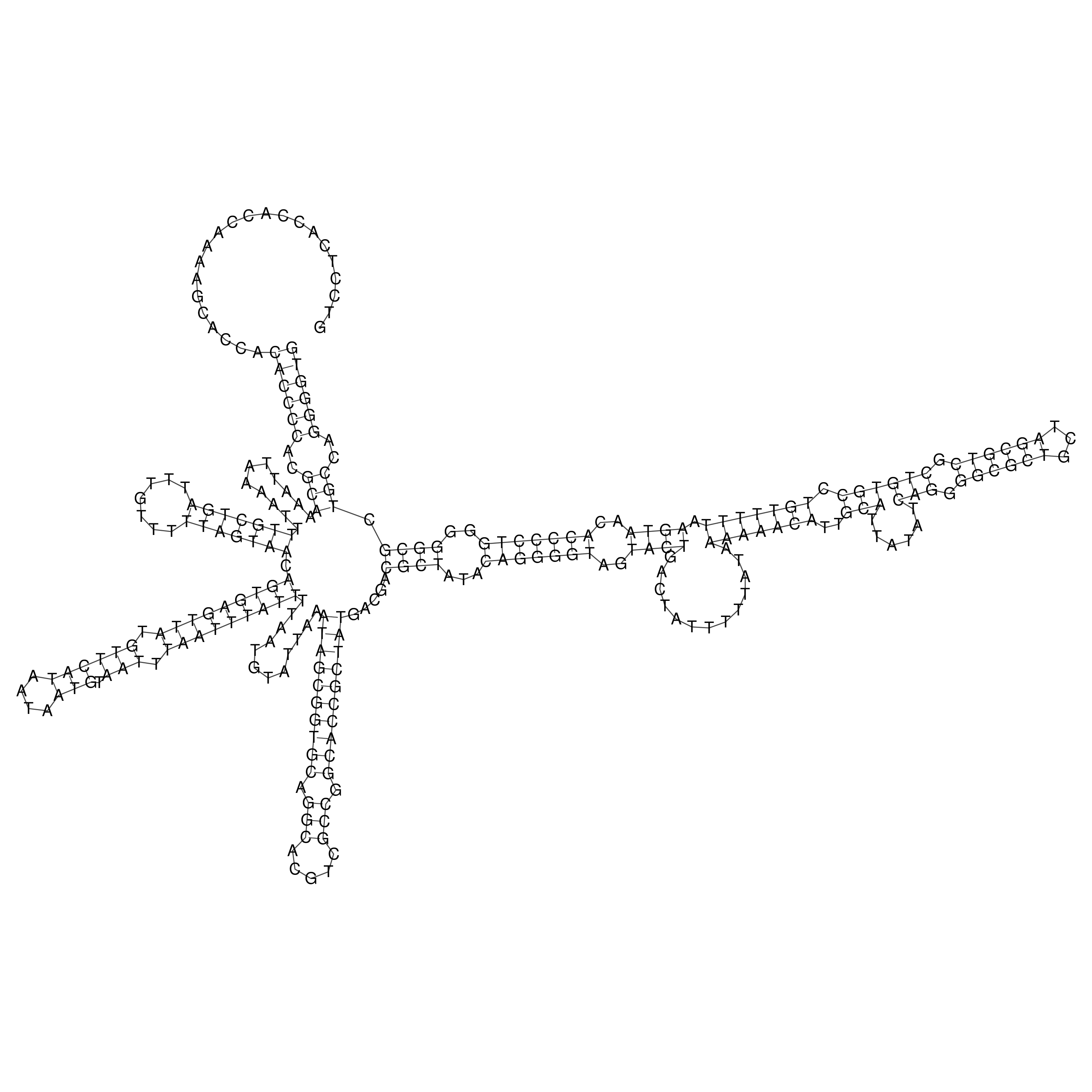
Relaxase
| ID | 2752 | GenBank | WP_024798951 |
| Name | traI_L1971_RS00420_pSCD1 |
UniProt ID | A0A379NF77 |
| Length | 1751 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1751 a.a. Molecular weight: 191019.00 Da Isoelectric Point: 5.8095
MMSIAQVRSAGSAAGYYSDRDNYYVLGSMEERWAGKGAEQLGLQGAVDKEVFTRVLEGRLPDGADLSRQQ
DGGNKHRPGYDLTFSAPKSVSLMAMLAGDKRLTEAHNQAVDIAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTLHDGEWKTLSSDKVGKTGFIENIYANQIAFGKIYRAVLKEKVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPAVKMAEWMQT
LKDTGFDISAYREAADRRAEIQAAQPVPSQEQPDIQQAVTQAIAGLSDRKVQFTYTDVLARTVGMLPPEA
GVIEKARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSSDIMKQNRVTVHPEKSVPRTGSYSD
AVSVLARDRPSLAIISGQGGAAGQRERVAELTMMAREQGREVQIIAADRRSQTNLKQDERLSGELITGRR
QLQEGMTFSPGSTVIVDQGEKLSLKETLTLLDGTARHNVQVLITDSGQRTGTGSALMAMKEAGVNSYRWQ
GGQQTPATVISEPDRNVRYARLAGDFVAAVKTGEESVAQVSGVREQAILAGMIRSELKTQGILGQQDTMM
TALSPVWLDSRNRYLRDMYREGMVMEQWNPEKRSHDRYVIDRVTAQSHSLTLRDAQGGTQMVRISALDSS
WSLFRPEKIPVADGERLMVTGKIPGLRVSGGDRLQVSAVNDGVMTVTVPGRAEPASLPVGDSPFTALKLE
NGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDEIRTAEKLSRHPSFTVVSEQIK
ARAGEVSLETAISLQKAGLHTPAQQALHLAIPVVESKNLAFSQVELLTEAKSFAAEGTGFADLGREIDAQ
IKRGDLLYVDVAKGYGTDLLISRASYDAEKSILHHILEGKEAVTPLMERVPGELMETLTSGQRAATRMIL
ETKDRFTVVQGYAGVGKTTQFRAVMSAVNLLPEDERPRVVGLGPTHRAVGEMRSAGVEAQTLASFLHDTQ
LQQRSGEAPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQARSAA
DVAIMKEIVRQTPELRDAVYSLINRDVNKALSGLENVKPVQVPRLKGAWAPENSVTEFSRLQERELAKAA
QEAEKKGEAFPDVPVTLYEAIVRDYTGRMPEAREQTLIVTHLNEDRRVLNSMIHDVREKAGELGEQQADI
PVLTTANIRDGELRRLSTWEAHPGALALVDNVYHRIAGISKEDGLITLEDKAGNTRLISPREAAAEGVTL
YNPETIRVGAGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGKQTCVVRPGQDRAEQHIDLAYAIT
AHGAQGASETFAIALEGTEGGRKQMAGFESAYVALSRMKQHVQVYTDDRQGWVKAINSAEQKGTAHDVLE
PKSEREMMNAERLFSTARELRDVAVGRAVLRNAGLAQGDSRARFIAPGRKYPQPYVALPAFDRNGKSAGI
WLNPLTTDDGAGLRGFTGEGRVKGSEEAQFVALQGSRNGESLLAGSMQDGVRIARENPDSGVVVRIAGDG
RPWNPVAITGGRVWGDIPDSNVQPGAGNGEPVTAEILAQRQAEEAVRRETERRADEIVRKMAEDKSDLPE
EKTAQAVREIAGQEQDRMTPPEREPALPESVLREPQRERETIREVARENRGRERLQEMVCDLQKEKTPGG
D
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1163 | GenBank | WP_001151546 |
| Name | WP_001151546_pSCD1 |
UniProt ID | Q93GP1 |
| Length | 126 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 126 a.a. Molecular weight: 14366.30 Da Isoelectric Point: 5.7236
MAKVNVYISNEVHNKITAIVEKRRQEGARDKDISFSGTSSMLLELGLRVYEAQMERKESPFNQTEFNKVL
LENVLKTQSSVAKILGIGSLSPHVAGNPKFEYANMVEDIKEKVSSEMERFFHENEE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q93GP1 |
| ID | 1164 | GenBank | WP_001676655 |
| Name | WP_001676655_pSCD1 |
UniProt ID | H3JT82 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9147.26 Da Isoelectric Point: 9.6183
MRRRNAREGISRTVSVYLDEDTNNRLIRAKDRSGRSKTIEVQIRLRDHLKRYPDFYNEEIFREVIEETES
TFKEI
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | H3JT82 |
T4CP
| ID | 2589 | GenBank | WP_024798976 |
| Name | traC_L1971_RS00310_pSCD1 |
UniProt ID | _ |
| Length | 882 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 882 a.a. Molecular weight: 100353.19 Da Isoelectric Point: 6.7137
MKNLLDTVTQAANSLLTALKMPDESAQANQILGEMSFPQFSRLLPYRDYNQESGLFMNDATMGFMLEATP
VNGANETIALSLDHLLRTKLPRGIPLCVHLMSSQLVGERIEYGLREFSWSGEQAERLNAITRAYYMKAAE
THFPLPEGMNLPLTLRHYRVFISYCAPSRKKSRADILEMENLVKIIRASLQGAYISTQSVDAKAFINIVG
EMINHNPESLHPTRRRLDPYSDLNYQCVDDSFDLKVRQDYLTLGLRDNGKNSTARIMSFHLARNPEIAFL
WNSADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGD
LRQRLASNQSSVVSYFFNITAFCRDNKEMALETEQDILNSFRKNGFELISPRFNHMRNFLACLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITNIDQSAERIRDQLSV
MASPNGNLDEVHEGLLLQAVRAAWLTKKNRARIDDVVDFLENARTSEQYAESPGIRSRLDEMIVLLNQYT
AAGTYGRYFNSDEPSLRDDARMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNRKVGDFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFPQLHREMIEKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLQEHETFRGTYEYKAETD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2590 | GenBank | WP_152280056 |
| Name | traD_L1971_RS00405_pSCD1 |
UniProt ID | _ |
| Length | 741 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 741 a.a. Molecular weight: 84003.85 Da Isoelectric Point: 5.2394
MSFNAKDMTQGGQIASMRIRMFSQIANIIFYCLFIFFWILVGLVLWVKLIWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIRYYGQTFRMNAAQVLQDKYTVWCGEQLWSAFVLAACVALVICLITFFMVTWILGRQGKQ
QSEDDVTGGRQLTDNPKDVARMLKKDGKASDILIGDLPIIKDSEIQNFLLHGTVSTGKSEIIRRLANYAR
QRGDMVVMYDRSCEFVKSYYDPSIDKILNPCDARCAAWDLWRECLTLPDFDNAANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDSDRSYSKLVDTLLSIKIEKLRTYLRDSPAANLVEEKIEKTAISIRAVLTNYVKA
VRYLQGIERNGEPFTIRDWMRGVREDQKNGWLFISSNADTHSSLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEIVPEARKFGGCYVFGIQSSTQMEDIYGEKAAATLLDVMNTRAFFRSPSYKIA
DYVAHEIGEKEILKASEQYSYGADPVRDGVSTGKEMERVTLVSYSDIQSLPDLTCYVTLPGPYPAVKLAL
KYQERPKVAPEFIPREMNPEAEARLSAVLAAREAEGRQMSSLFEPDTEAVAPAAAEAQAEQPQQPQQPQQ
PQQPQQPQQPQQPQQPQSTVVSDKKSAAAGTAVNTPAGGVGQELKMKQEDEEQPLPPGINASGEVVDMAV
YERWRQEQELNTQQQMQRREEVNINVHRGHGKDEPEPGDHF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 38485..66337
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| L1971_RS00205 (L1971_00205) | 33604..34026 | - | 423 | WP_000925628 | translesion error-prone DNA polymerase V autoproteolytic subunit | - |
| L1971_RS00210 (L1971_00210) | 34327..36048 | + | 1722 | Protein_41 | ParB/RepB/Spo0J family partition protein | - |
| L1971_RS00215 (L1971_00215) | 36088..36189 | + | 102 | Protein_42 | conjugation system SOS inhibitor PsiB family protein | - |
| L1971_RS00220 (L1971_00220) | 36190..36846 | + | 657 | Protein_43 | plasmid SOS inhibition protein A | - |
| L1971_RS23195 | 36881..37009 | + | 129 | WP_000372430 | hypothetical protein | - |
| L1971_RS00225 (L1971_00225) | 37095..37253 | + | 159 | WP_001704449 | hypothetical protein | - |
| L1971_RS00230 (L1971_00230) | 37356..38174 | + | 819 | Protein_46 | DUF932 domain-containing protein | - |
| L1971_RS00235 (L1971_00235) | 38485..38955 | - | 471 | WP_015056628 | transglycosylase SLT domain-containing protein | virB1 |
| L1971_RS00240 (L1971_00240) | 39370..39750 | + | 381 | WP_001151546 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| L1971_RS00245 (L1971_00245) | 39943..40629 | + | 687 | WP_000336296 | PAS domain-containing protein | - |
| L1971_RS00250 (L1971_00250) | 40723..40950 | + | 228 | WP_001676655 | conjugal transfer relaxosome protein TraY | - |
| L1971_RS00255 (L1971_00255) | 40992..41354 | + | 363 | WP_001274198 | type IV conjugative transfer system pilin TraA | - |
| L1971_RS00260 (L1971_00260) | 41369..41680 | + | 312 | WP_000012129 | type IV conjugative transfer system protein TraL | traL |
| L1971_RS00265 (L1971_00265) | 41702..42268 | + | 567 | WP_000399761 | type IV conjugative transfer system protein TraE | traE |
| L1971_RS00270 (L1971_00270) | 42255..42995 | + | 741 | WP_001230721 | type-F conjugative transfer system secretin TraK | traK |
| L1971_RS00275 (L1971_00275) | 42995..44416 | + | 1422 | WP_000146702 | F-type conjugal transfer pilus assembly protein TraB | traB |
| L1971_RS00280 (L1971_00280) | 44406..45002 | + | 597 | WP_024799158 | conjugal transfer pilus-stabilizing protein TraP | - |
| L1971_RS00285 (L1971_00285) | 44980..45204 | + | 225 | WP_077906918 | DUF2689 domain-containing protein | - |
| L1971_RS00290 (L1971_00290) | 45201..45716 | + | 516 | WP_001079997 | type IV conjugative transfer system lipoprotein TraV | traV |
| L1971_RS00295 (L1971_00295) | 45851..46072 | + | 222 | WP_001278681 | conjugal transfer protein TraR | - |
| L1971_RS00300 (L1971_00300) | 46065..46532 | + | 468 | WP_000549560 | hypothetical protein | - |
| L1971_RS00305 (L1971_00305) | 46529..46912 | + | 384 | WP_000870324 | hypothetical protein | - |
| L1971_RS00310 (L1971_00310) | 46928..49576 | + | 2649 | WP_024798976 | type IV secretion system protein TraC | virb4 |
| L1971_RS00315 (L1971_00315) | 49551..49937 | + | 387 | WP_000105164 | type-F conjugative transfer system protein TrbI | - |
| L1971_RS00320 (L1971_00320) | 49934..50566 | + | 633 | WP_000871997 | type-F conjugative transfer system protein TraW | traW |
| L1971_RS00325 (L1971_00325) | 50563..51555 | + | 993 | WP_000833321 | conjugal transfer pilus assembly protein TraU | traU |
| L1971_RS00330 (L1971_00330) | 51574..51870 | + | 297 | WP_077914412 | hypothetical protein | - |
| L1971_RS00335 (L1971_00335) | 51867..52496 | + | 630 | WP_000089759 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| L1971_RS00340 (L1971_00340) | 52517..52654 | - | 138 | WP_154071808 | hypothetical protein | - |
| L1971_RS00345 (L1971_00345) | 52856..54733 | + | 1878 | WP_237123561 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| L1971_RS00350 (L1971_00350) | 54744..55220 | + | 477 | WP_237123570 | hypothetical protein | - |
| L1971_RS00355 (L1971_00355) | 55230..55445 | + | 216 | WP_000783297 | conjugal transfer protein TrbE | - |
| L1971_RS00360 (L1971_00360) | 55457..56206 | + | 750 | WP_237123651 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| L1971_RS00365 (L1971_00365) | 56221..56565 | + | 345 | WP_001697170 | conjugal transfer protein TrbA | - |
| L1971_RS00370 (L1971_00370) | 56941..57234 | + | 294 | WP_001233880 | type-F conjugative transfer system pilin chaperone TraQ | - |
| L1971_RS00375 (L1971_00375) | 57221..57769 | + | 549 | WP_024798870 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| L1971_RS00380 (L1971_00380) | 57759..59141 | + | 1383 | WP_237123578 | conjugal transfer pilus assembly protein TraH | traH |
| L1971_RS00385 (L1971_00385) | 59138..61957 | + | 2820 | WP_024798871 | conjugal transfer mating-pair stabilization protein TraG | traG |
| L1971_RS00390 (L1971_00390) | 61960..62271 | - | 312 | WP_000522257 | hypothetical protein | - |
| L1971_RS00395 (L1971_00395) | 62370..63101 | + | 732 | WP_000782437 | conjugal transfer complement resistance protein TraT | - |
| L1971_RS00400 (L1971_00400) | 63353..63832 | + | 480 | WP_025649166 | hypothetical protein | - |
| L1971_RS00405 (L1971_00405) | 64112..66337 | + | 2226 | WP_152280056 | type IV conjugative transfer system coupling protein TraD | virb4 |
| L1971_RS00410 (L1971_00410) | 66346..66744 | - | 399 | WP_000911323 | tRNA(fMet)-specific endonuclease VapC | - |
| L1971_RS00415 (L1971_00415) | 66744..66971 | - | 228 | WP_000450263 | toxin-antitoxin system antitoxin VapB | - |
Host bacterium
| ID | 4157 | GenBank | NZ_CP091299 |
| Plasmid name | pSCD1 | Incompatibility group | IncFII |
| Plasmid size | 104427 bp | Coordinate of oriT [Strand] | 39026..39295 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar Bovismorbificans strain SB699 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | spvC, spvB, rck, pefD, pefC, pefA, pefB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |