Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103708 |
| Name | oriT_JS187-vir|unnamed3 |
| Organism | Klebsiella pneumoniae strain JS187-vir |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MZ475702 (71725..71774 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_JS187-vir|unnamed3
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file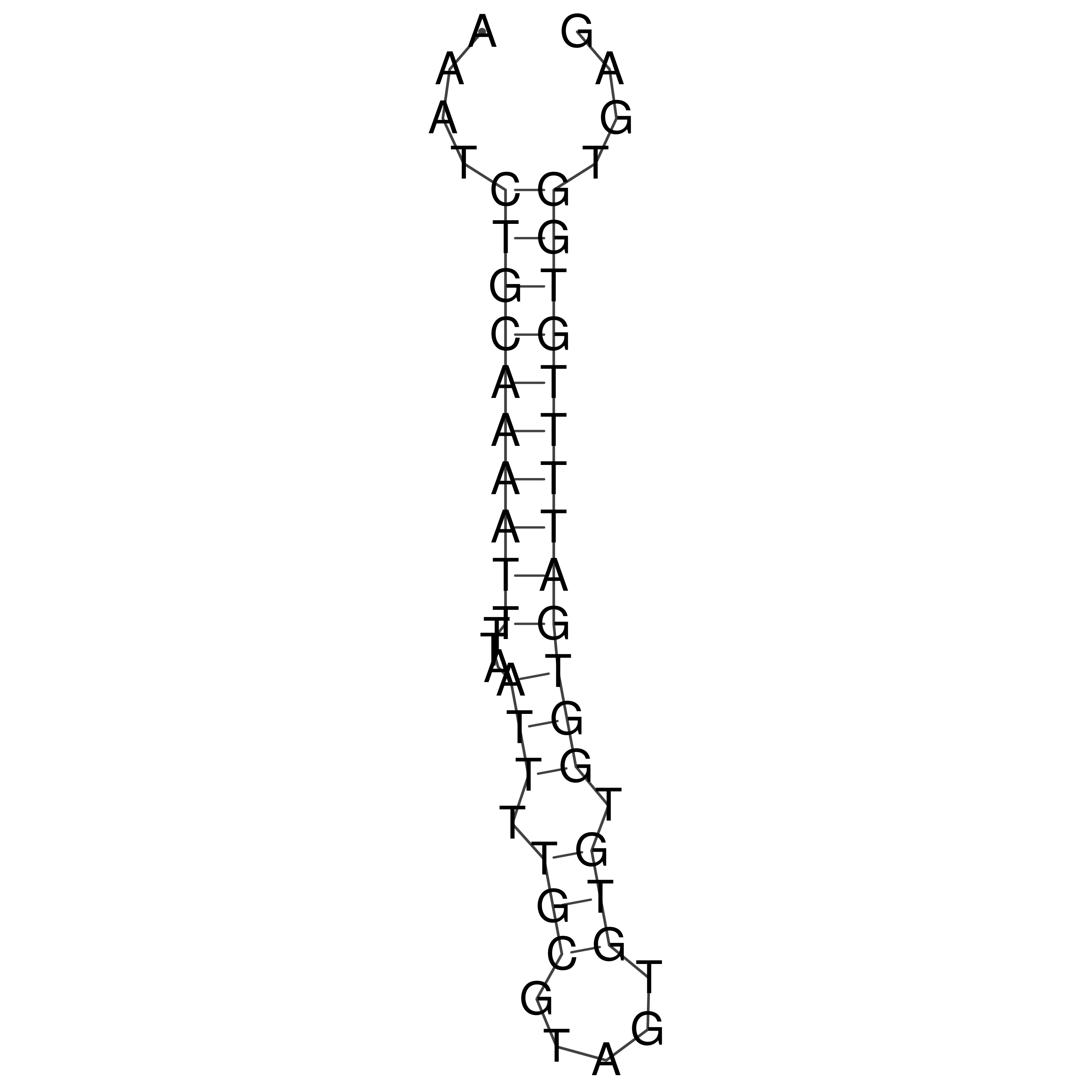
Relaxase
| ID | 2749 | GenBank | WP_014343483 |
| Name | traI_LL121_RS00645_JS187-vir|unnamed3 |
UniProt ID | A0A0H3H1C0 |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190459.18 Da Isoelectric Point: 6.1353
>WP_014343483.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREATDARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGINAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVTF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRASTRM
ILETPDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPHVIGLGPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSISPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDLHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREATDARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGINAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVTF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRASTRM
ILETPDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPHVIGLGPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSISPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDLHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A0H3H1C0 |
T4CP
| ID | 2586 | GenBank | WP_110133658 |
| Name | traD_LL121_RS00640_JS187-vir|unnamed3 |
UniProt ID | _ |
| Length | 760 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 760 a.a. Molecular weight: 84917.94 Da Isoelectric Point: 5.0045
>WP_110133658.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSPPA
PAEMTVSPAPVKAPPTTKRPAAEPPVLPVTPIPLLKPKAAAAAATASSAGTPAAAAGGTEQELAQQSAEQ
GQDMLPAGMNEDGVIEDMQAYDAWLADDQTQRDMQRREEVNINHSHRHDEQDDVEIGGNI
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSPPA
PAEMTVSPAPVKAPPTTKRPAAEPPVLPVTPIPLLKPKAAAAAATASSAGTPAAAAGGTEQELAQQSAEQ
GQDMLPAGMNEDGVIEDMQAYDAWLADDQTQRDMQRREEVNINHSHRHDEQDDVEIGGNI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 71167..99622
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LL121_RS00430 (MGOKJAAN_00085) | 66323..66640 | + | 318 | WP_014343505 | hypothetical protein | - |
| LL121_RS00435 (MGOKJAAN_00086) | 67481..67837 | + | 357 | WP_014343501 | hypothetical protein | - |
| LL121_RS00440 (MGOKJAAN_00087) | 67898..68110 | + | 213 | WP_014343500 | hypothetical protein | - |
| LL121_RS00445 (MGOKJAAN_00088) | 68121..68345 | + | 225 | WP_014343499 | hypothetical protein | - |
| LL121_RS00450 (MGOKJAAN_00089) | 68426..68746 | + | 321 | WP_014343498 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| LL121_RS00455 (MGOKJAAN_00090) | 68736..69014 | + | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| LL121_RS00460 (MGOKJAAN_00091) | 69018..69122 | + | 105 | Protein_91 | transcriptional regulator | - |
| LL121_RS00465 (MGOKJAAN_00092) | 69951..70772 | + | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| LL121_RS00470 (MGOKJAAN_00093) | 70805..71134 | + | 330 | WP_013214015 | DUF5983 family protein | - |
| LL121_RS00475 (MGOKJAAN_00094) | 71167..71652 | - | 486 | WP_014343495 | transglycosylase SLT domain-containing protein | virB1 |
| LL121_RS00480 (MGOKJAAN_00095) | 72043..72459 | + | 417 | WP_014343494 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| LL121_RS00485 (MGOKJAAN_00096) | 72659..73396 | + | 738 | WP_013214018 | conjugal transfer protein TrbJ | - |
| LL121_RS00490 | 73511..73717 | + | 207 | WP_171773970 | TraY domain-containing protein | - |
| LL121_RS00495 (MGOKJAAN_00097) | 73779..74147 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| LL121_RS00500 (MGOKJAAN_00098) | 74161..74466 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| LL121_RS00505 (MGOKJAAN_00099) | 74486..75052 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| LL121_RS00510 (MGOKJAAN_00100) | 75039..75779 | + | 741 | WP_013214019 | type-F conjugative transfer system secretin TraK | traK |
| LL121_RS00515 (MGOKJAAN_00101) | 75779..77203 | + | 1425 | WP_013214020 | F-type conjugal transfer pilus assembly protein TraB | traB |
| LL121_RS00520 (MGOKJAAN_00102) | 77275..77859 | + | 585 | WP_014343493 | type IV conjugative transfer system lipoprotein TraV | traV |
| LL121_RS00525 (MGOKJAAN_00103) | 78182..78400 | + | 219 | WP_004195468 | hypothetical protein | - |
| LL121_RS00530 (MGOKJAAN_00104) | 78401..78712 | + | 312 | WP_013214022 | hypothetical protein | - |
| LL121_RS00535 (MGOKJAAN_00105) | 78779..79183 | + | 405 | WP_004197817 | hypothetical protein | - |
| LL121_RS00540 (MGOKJAAN_00107) | 79560..79958 | + | 399 | WP_014343490 | hypothetical protein | - |
| LL121_RS00545 (MGOKJAAN_00108) | 80030..82669 | + | 2640 | WP_013214024 | type IV secretion system protein TraC | virb4 |
| LL121_RS00550 (MGOKJAAN_00109) | 82669..83058 | + | 390 | WP_013214025 | type-F conjugative transfer system protein TrbI | - |
| LL121_RS00555 (MGOKJAAN_00110) | 83058..83693 | + | 636 | WP_013214026 | type-F conjugative transfer system protein TraW | traW |
| LL121_RS00560 (MGOKJAAN_00111) | 83728..84129 | + | 402 | WP_004194979 | hypothetical protein | - |
| LL121_RS00565 (MGOKJAAN_00112) | 84126..85115 | + | 990 | WP_013214027 | conjugal transfer pilus assembly protein TraU | traU |
| LL121_RS00570 (MGOKJAAN_00113) | 85128..85766 | + | 639 | WP_004193871 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LL121_RS00575 (MGOKJAAN_00114) | 85825..87780 | + | 1956 | WP_014343489 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| LL121_RS00580 (MGOKJAAN_00115) | 87812..88066 | + | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| LL121_RS00585 (MGOKJAAN_00116) | 88044..88292 | + | 249 | WP_013214029 | hypothetical protein | - |
| LL121_RS00590 (MGOKJAAN_00117) | 88305..88631 | + | 327 | WP_013214030 | hypothetical protein | - |
| LL121_RS00595 (MGOKJAAN_00118) | 88652..89404 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| LL121_RS00600 (MGOKJAAN_00119) | 89415..89654 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| LL121_RS00605 (MGOKJAAN_00120) | 89626..90183 | + | 558 | WP_013214031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| LL121_RS00610 (MGOKJAAN_00121) | 90229..90672 | + | 444 | WP_014343488 | F-type conjugal transfer protein TrbF | - |
| LL121_RS00615 (MGOKJAAN_00122) | 90650..92029 | + | 1380 | WP_014343487 | conjugal transfer pilus assembly protein TraH | traH |
| LL121_RS00620 (MGOKJAAN_00123) | 92029..94878 | + | 2850 | WP_013609534 | conjugal transfer mating-pair stabilization protein TraG | traG |
| LL121_RS00625 (MGOKJAAN_00124) | 94881..95414 | + | 534 | WP_014343486 | conjugal transfer protein TraS | - |
| LL121_RS00630 (MGOKJAAN_00125) | 95600..96331 | + | 732 | WP_013023830 | conjugal transfer complement resistance protein TraT | - |
| LL121_RS00635 (MGOKJAAN_00126) | 96524..97213 | + | 690 | WP_014343485 | hypothetical protein | - |
| LL121_RS00640 (MGOKJAAN_00127) | 97340..99622 | + | 2283 | WP_110133658 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 4151 | GenBank | NZ_MZ475702 |
| Plasmid name | JS187-vir|unnamed3 | Incompatibility group | IncFII |
| Plasmid size | 110080 bp | Coordinate of oriT [Strand] | 71725..71774 [-] |
| Host baterium | Klebsiella pneumoniae strain JS187-vir |
Cargo genes
| Drug resistance gene | blaKPC-2, blaTEM-1B |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |