Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103704 |
| Name | oriT_KP36M|unnamed1 |
| Organism | Klebsiella pneumoniae strain KP36M |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP017778 (184672..184822 [-], 151 nt) |
| oriT length | 151 nt |
| IRs (inverted repeats) | 93..98, 106..111 (TGGCCT..AGGCCA) 12..17, 24..29 (AACCCT..AGGGTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 151 nt
>oriT_KP36M|unnamed1
TGGTCGATGTGAACCCTTTCGACAGGGTTATGAATGAATTGAAAAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAA
TGGTCGATGTGAACCCTTTCGACAGGGTTATGAATGAATTGAAAAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file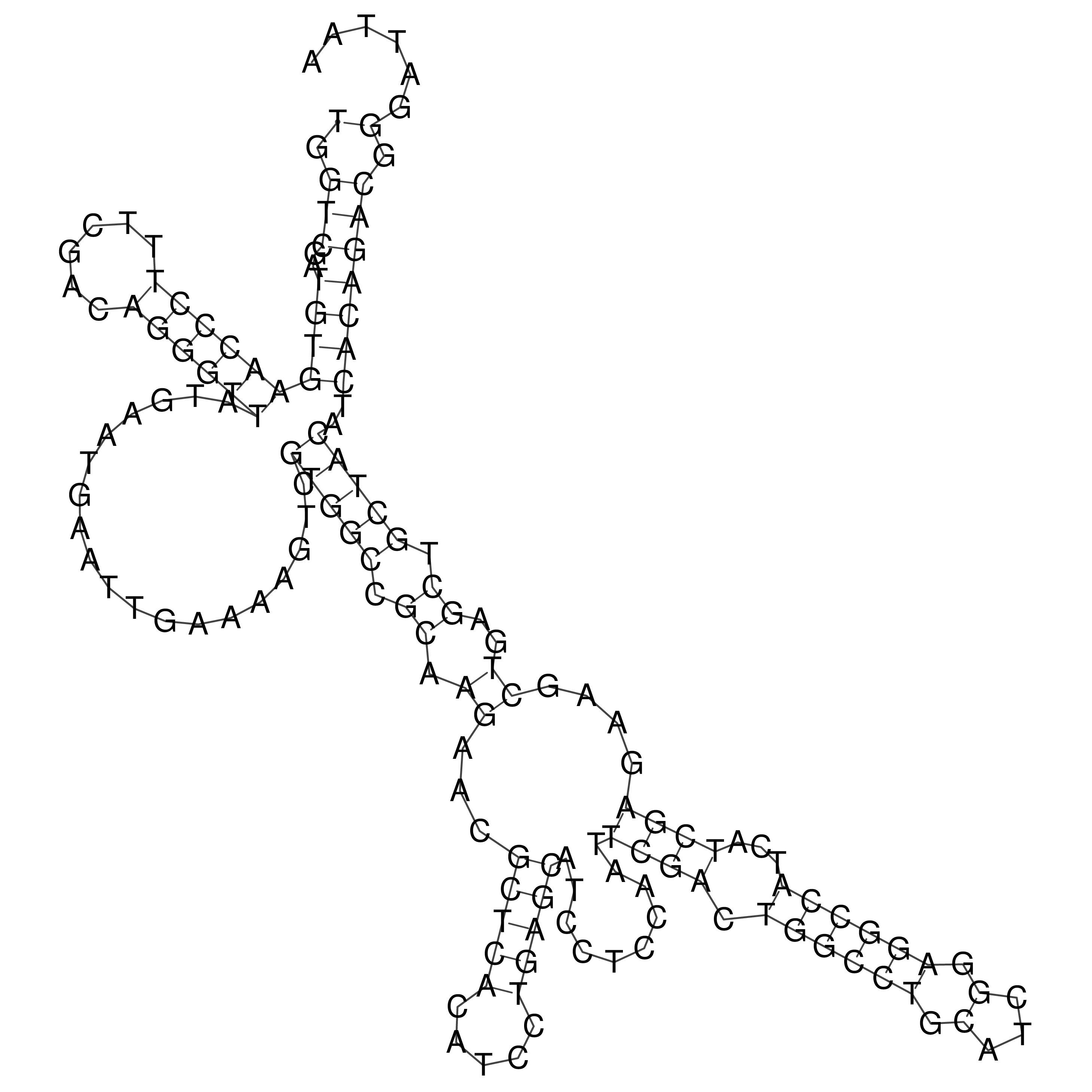
Relaxase
| ID | 2748 | GenBank | WP_001337757 |
| Name | mobH_BK002_RS26845_KP36M|unnamed1 |
UniProt ID | A0A709FRI2 |
| Length | 992 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 992 a.a. Molecular weight: 109581.85 Da Isoelectric Point: 4.8956
>WP_001337757.1 MULTISPECIES: MobH family relaxase [Gammaproteobacteria]
MLKALNKLFGGRSGVIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEADDGDDQEEGEAALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGPTESSK
PDAGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPF
DAFNASAETTSTDATNSEIPDVAMPGKQEEQPKQDFVPQEQNSLQGDDFPMFGGSDEPPSWAIEPLPMLT
DAPEQPTHTPEMPHTDNVNQHEKDAKTLLVEMLSGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVI
LYPDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQ
DAFELVSPPGLDASKNKSAPKQQSRKKAQQQKPEVNAGKASPEQKAKGKDSQPQPKEKKVDVTSPVEEQQ
RKPVQEKQNVARLPKREAQPVAPEPKVEREKELGHVEVREREDPEVREFEPPKAKTNPKDINAEDFLPSG
VTPQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKK
RQGKLYLEVNET
MLKALNKLFGGRSGVIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEADDGDDQEEGEAALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGPTESSK
PDAGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPF
DAFNASAETTSTDATNSEIPDVAMPGKQEEQPKQDFVPQEQNSLQGDDFPMFGGSDEPPSWAIEPLPMLT
DAPEQPTHTPEMPHTDNVNQHEKDAKTLLVEMLSGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVI
LYPDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQ
DAFELVSPPGLDASKNKSAPKQQSRKKAQQQKPEVNAGKASPEQKAKGKDSQPQPKEKKVDVTSPVEEQQ
RKPVQEKQNVARLPKREAQPVAPEPKVEREKELGHVEVREREDPEVREFEPPKAKTNPKDINAEDFLPSG
VTPQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKK
RQGKLYLEVNET
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A709FRI2 |
T4CP
| ID | 2584 | GenBank | WP_000637386 |
| Name | traC_BK002_RS26740_KP36M|unnamed1 |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 92590.87 Da Isoelectric Point: 5.9222
>WP_000637386.1 MULTISPECIES: type IV secretion system protein TraC [Gammaproteobacteria]
MIVTIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSAGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTTLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTERIFAKTNKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQQLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSVFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRQQAA
MIVTIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSAGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTTLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTERIFAKTNKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQQLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSVFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRQQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2585 | GenBank | WP_000178856 |
| Name | traD_BK002_RS26840_KP36M|unnamed1 |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 621 a.a. Molecular weight: 69197.66 Da Isoelectric Point: 6.9403
>WP_000178856.1 MULTISPECIES: conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPDDMWLGSGFLWENRHAQRVFEILKRDWTSIVGRESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVMPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPDDMWLGSGFLWENRHAQRVFEILKRDWTSIVGRESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVMPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 177648..186738
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BK002_RS26770 (BK002_27235) | 172917..176174 | + | 3258 | WP_004152343 | type I restriction endonuclease | - |
| BK002_RS26775 (BK002_27240) | 176179..177447 | + | 1269 | WP_004152342 | ISL3-like element ISKpn25 family transposase | - |
| BK002_RS26780 (BK002_27245) | 177648..178226 | - | 579 | WP_000793435 | type IV conjugative transfer system lipoprotein TraV | traV |
| BK002_RS26785 (BK002_27250) | 178223..179536 | - | 1314 | WP_024131605 | TraB/VirB10 family protein | traB |
| BK002_RS26790 (BK002_27255) | 179536..180453 | - | 918 | WP_000794249 | type-F conjugative transfer system secretin TraK | traK |
| BK002_RS26795 (BK002_27260) | 180437..181063 | - | 627 | WP_001049716 | TraE/TraK family type IV conjugative transfer system protein | traE |
| BK002_RS26800 (BK002_27265) | 181060..181341 | - | 282 | WP_000805625 | type IV conjugative transfer system protein TraL | traL |
| BK002_RS26805 (BK002_27270) | 181486..181851 | - | 366 | WP_001052531 | hypothetical protein | - |
| BK002_RS26810 | 181863..182006 | - | 144 | WP_001275801 | hypothetical protein | - |
| BK002_RS26815 (BK002_27275) | 182187..182849 | - | 663 | WP_001231463 | hypothetical protein | - |
| BK002_RS26820 (BK002_27280) | 182849..183226 | - | 378 | WP_000869261 | hypothetical protein | - |
| BK002_RS26825 (BK002_27285) | 183236..183682 | - | 447 | WP_000122506 | hypothetical protein | - |
| BK002_RS26830 (BK002_27290) | 183692..184321 | - | 630 | WP_000743450 | DUF4400 domain-containing protein | tfc7 |
| BK002_RS26835 (BK002_27295) | 184278..184823 | - | 546 | WP_000228720 | hypothetical protein | - |
| BK002_RS26840 (BK002_27300) | 184873..186738 | - | 1866 | WP_000178856 | conjugative transfer system coupling protein TraD | virb4 |
| BK002_RS26845 (BK002_27305) | 186735..189713 | - | 2979 | WP_001337757 | MobH family relaxase | - |
| BK002_RS26850 (BK002_27310) | 189881..190498 | + | 618 | WP_001249396 | hypothetical protein | - |
| BK002_RS26855 (BK002_27315) | 190480..190713 | + | 234 | WP_001191892 | hypothetical protein | - |
Host bacterium
| ID | 4147 | GenBank | NZ_CP017778 |
| Plasmid name | KP36M|unnamed1 | Incompatibility group | IncFIB |
| Plasmid size | 225961 bp | Coordinate of oriT [Strand] | 184672..184822 [-] |
| Host baterium | Klebsiella pneumoniae strain KP36M |
Cargo genes
| Drug resistance gene | sul1, qnrB2, sul2, erm(42) |
| Virulence gene | - |
| Metal resistance gene | merR, merT, merP, merC, merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |