Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103646 |
| Name | oriT_pKPC-063001 |
| Organism | Klebsiella pneumoniae strain KP63 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MZ156798 (195821..195870 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pKPC-063001
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTCATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTCATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file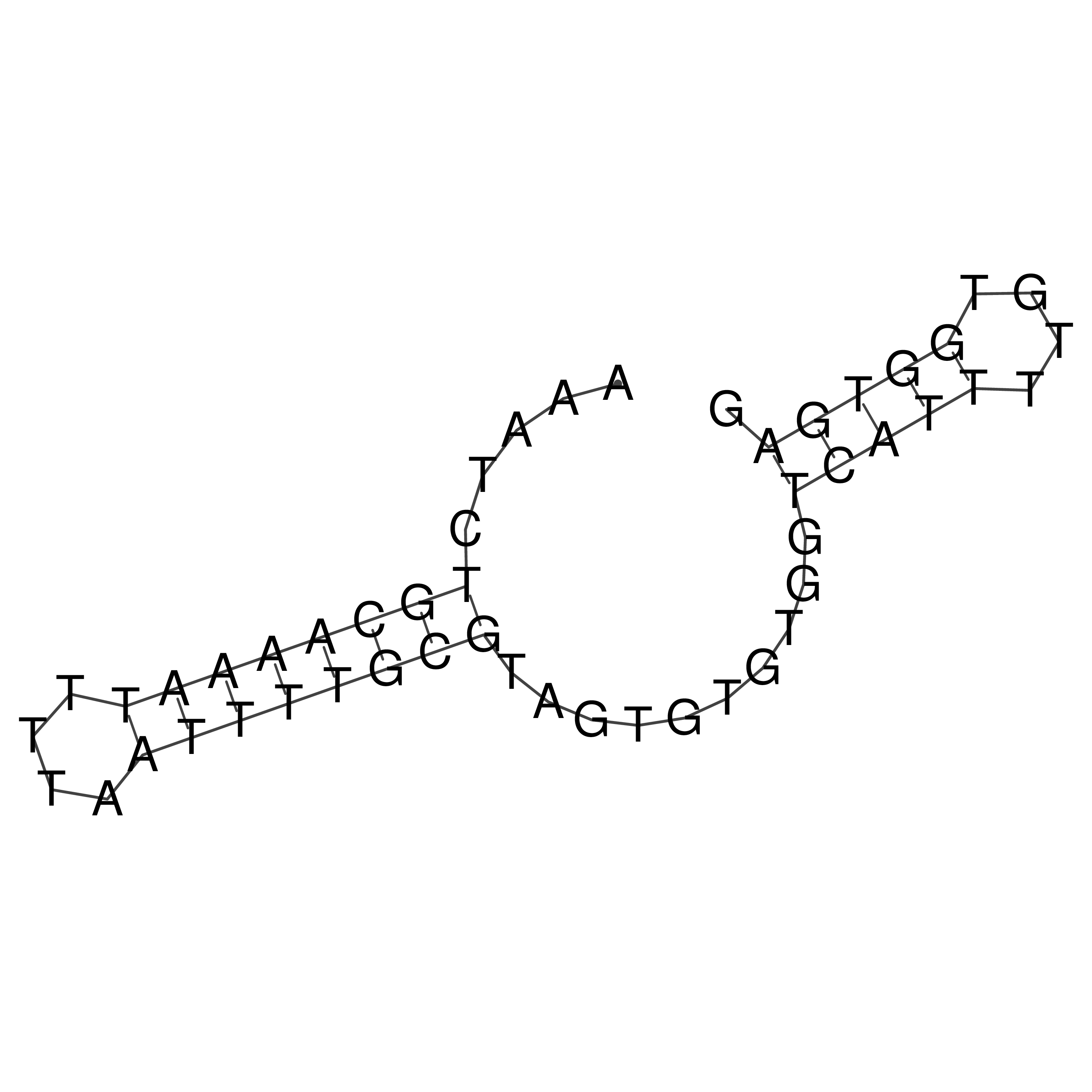
Relaxase
| ID | 2716 | GenBank | WP_032448280 |
| Name | traI_KR465_RS00815_pKPC-063001 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190581.22 Da Isoelectric Point: 5.9995
>WP_032448280.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWLTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWLTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2544 | GenBank | WP_013214038 |
| Name | traD_KR465_RS00820_pKPC-063001 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 86010.07 Da Isoelectric Point: 5.1787
>WP_013214038.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 166409..196428
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KR465_RS00820 | 166409..168721 | - | 2313 | WP_013214038 | type IV conjugative transfer system coupling protein TraD | virb4 |
| KR465_RS00825 | 168848..169537 | - | 690 | WP_014343485 | hypothetical protein | - |
| KR465_RS00830 | 169730..170461 | - | 732 | WP_013214036 | conjugal transfer complement resistance protein TraT | - |
| KR465_RS00835 | 170713..171315 | - | 603 | WP_013214035 | conjugal transfer protein TraS | - |
| KR465_RS00840 | 171338..174160 | - | 2823 | WP_013214034 | conjugal transfer mating-pair stabilization protein TraG | traG |
| KR465_RS00845 | 174160..175539 | - | 1380 | WP_074160414 | conjugal transfer pilus assembly protein TraH | traH |
| KR465_RS00850 | 175517..175960 | - | 444 | WP_013214032 | F-type conjugal transfer protein TrbF | - |
| KR465_RS00855 | 176006..176563 | - | 558 | WP_013214031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| KR465_RS00860 | 176535..176774 | - | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| KR465_RS00865 | 176785..177537 | - | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| KR465_RS00870 | 177558..177884 | - | 327 | WP_013214030 | hypothetical protein | - |
| KR465_RS00875 | 177897..178145 | - | 249 | WP_013214029 | hypothetical protein | - |
| KR465_RS00880 | 178123..178377 | - | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| KR465_RS00885 | 178409..180364 | - | 1956 | WP_032448279 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| KR465_RS00890 | 180423..181070 | - | 648 | WP_015632510 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| KR465_RS00895 | 181149..181325 | - | 177 | WP_162866177 | hypothetical protein | - |
| KR465_RS00900 | 181346..182335 | - | 990 | WP_032439620 | conjugal transfer pilus assembly protein TraU | traU |
| KR465_RS00905 | 182332..182733 | - | 402 | WP_016831047 | hypothetical protein | - |
| KR465_RS00910 | 182768..183403 | - | 636 | WP_020325113 | type-F conjugative transfer system protein TraW | traW |
| KR465_RS00915 | 183403..183792 | - | 390 | WP_004167468 | type-F conjugative transfer system protein TrbI | - |
| KR465_RS00920 | 183792..186431 | - | 2640 | WP_032448278 | type IV secretion system protein TraC | virb4 |
| KR465_RS00925 | 186503..186901 | - | 399 | WP_023158008 | hypothetical protein | - |
| KR465_RS00930 | 186909..187298 | - | 390 | WP_065799586 | hypothetical protein | - |
| KR465_RS00935 | 187341..187745 | - | 405 | WP_016831042 | hypothetical protein | - |
| KR465_RS00940 | 187812..188124 | - | 313 | Protein_191 | hypothetical protein | - |
| KR465_RS00945 | 188125..188343 | - | 219 | WP_014386199 | hypothetical protein | - |
| KR465_RS00950 | 188367..188657 | - | 291 | WP_065799585 | hypothetical protein | - |
| KR465_RS00955 | 188662..189072 | - | 411 | WP_014386197 | hypothetical protein | - |
| KR465_RS00960 | 189203..189787 | - | 585 | WP_014386196 | type IV conjugative transfer system lipoprotein TraV | traV |
| KR465_RS00965 | 189834..190406 | - | 573 | WP_014386195 | conjugal transfer pilus-stabilizing protein TraP | - |
| KR465_RS00970 | 190399..191823 | - | 1425 | WP_014386194 | F-type conjugal transfer pilus assembly protein TraB | traB |
| KR465_RS00975 | 191823..192563 | - | 741 | WP_014386193 | type-F conjugative transfer system secretin TraK | traK |
| KR465_RS00980 | 192550..193116 | - | 567 | WP_004152602 | type IV conjugative transfer system protein TraE | traE |
| KR465_RS00985 | 193136..193441 | - | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| KR465_RS00990 | 193455..193823 | - | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| KR465_RS00995 | 193885..194091 | - | 207 | WP_171773970 | TraY domain-containing protein | - |
| KR465_RS01000 | 194224..194943 | - | 720 | WP_014386192 | conjugal transfer protein | - |
| KR465_RS01005 | 195136..195552 | - | 417 | WP_072145360 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| KR465_RS01010 | 195943..196428 | + | 486 | WP_004178063 | transglycosylase SLT domain-containing protein | virB1 |
| KR465_RS01015 | 196461..196790 | - | 330 | WP_014386191 | DUF5983 family protein | - |
| KR465_RS01020 | 196823..197644 | - | 822 | WP_004182076 | DUF932 domain-containing protein | - |
| KR465_RS01025 | 198477..198890 | - | 414 | WP_013023817 | helix-turn-helix domain-containing protein | - |
| KR465_RS01030 | 198891..199169 | - | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| KR465_RS01035 | 199159..199479 | - | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| KR465_RS01040 | 199560..199784 | - | 225 | WP_014386189 | hypothetical protein | - |
| KR465_RS01045 | 199795..200007 | - | 213 | WP_014386188 | hypothetical protein | - |
| KR465_RS01050 | 200069..200395 | - | 327 | WP_014386187 | hypothetical protein | - |
Host bacterium
| ID | 4089 | GenBank | NZ_MZ156798 |
| Plasmid name | pKPC-063001 | Incompatibility group | IncFII |
| Plasmid size | 359625 bp | Coordinate of oriT [Strand] | 195821..195870 [+] |
| Host baterium | Klebsiella pneumoniae strain KP63 |
Cargo genes
| Drug resistance gene | blaKPC-2 |
| Virulence gene | iutA, iucD, iucC, iucB, iucA |
| Metal resistance gene | silE, silS, silR, silC, silF, silB, silA, silP, pcoA, pcoB, pcoC, pcoD, pcoR, pcoS, pcoE, arsR, arsD, arsA, arsB, arsC, fecD, fecE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |