Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103642 |
| Name | oriT_pDM86-optrA |
| Organism | Enterococcus faecalis strain DM86 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP116964 (36872..36912 [+], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 14..22, 29..37 (TAAAGTATA..TATACTTTA) 2..8, 13..19 (ACTTTAT..ATAAAGT) |
| Location of nic site | 27..28 |
| Conserved sequence flanking the nic site |
GTGTGTTATA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pDM86-optrA
CACTTTATGAATATAAAGTATAGTGTGTTATACTTTACATG
CACTTTATGAATATAAAGTATAGTGTGTTATACTTTACATG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file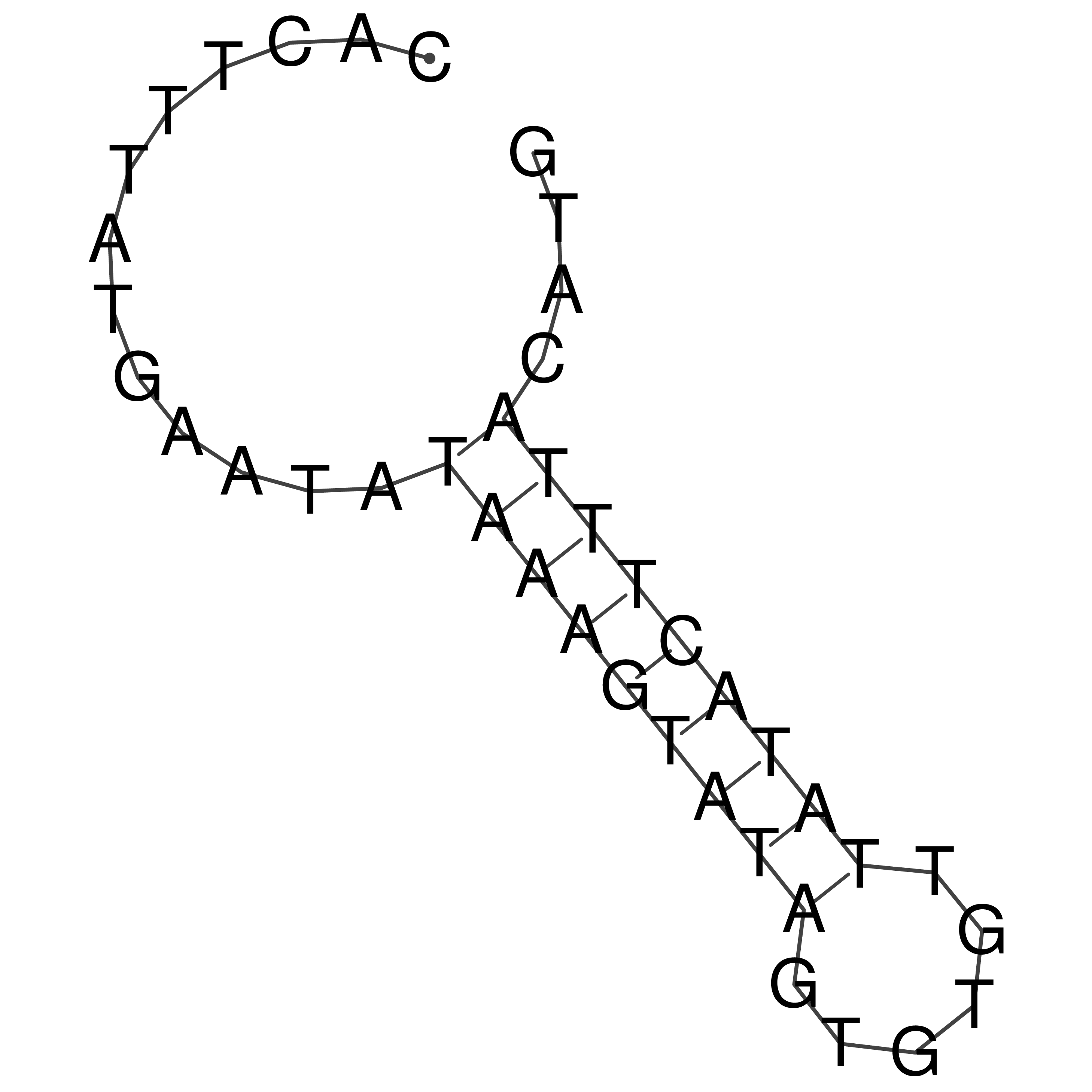
Relaxase
| ID | 2713 | GenBank | WP_272731721 |
| Name | mobV_PO880_RS15035_pDM86-optrA |
UniProt ID | _ |
| Length | 306 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 306 a.a. Molecular weight: 36017.83 Da Isoelectric Point: 6.1112
>WP_272731721.1 MobV family relaxase [Enterococcus faecalis]
MSMIVARMQKMKAENLVGIGNHNQRKTKNHSNPDIDTSLSKLNYDLVDRTQNYKTDIENFINENKSTTRA
VRKDAVLVNEWIISSDKDFFDNLTESEIENFFERSKDYFAEKFGEKNIRYATVHLDESTPHMHMGIVPFD
KDNKLSAKRVFNRQALRDVQEELPKYLQDFQFEIARGQKGSERKNLTVPEFKKLKEEELEIKKELQIKKD
ELIAYTKENQIDKKLDITPIKEMEDIEIETDEKTLFGKNKTEIVRQWTGNIILSENDYLKLNKEIKKGKK
TEGRLAAILETDVYQENKELKNELKD
MSMIVARMQKMKAENLVGIGNHNQRKTKNHSNPDIDTSLSKLNYDLVDRTQNYKTDIENFINENKSTTRA
VRKDAVLVNEWIISSDKDFFDNLTESEIENFFERSKDYFAEKFGEKNIRYATVHLDESTPHMHMGIVPFD
KDNKLSAKRVFNRQALRDVQEELPKYLQDFQFEIARGQKGSERKNLTVPEFKKLKEEELEIKKELQIKKD
ELIAYTKENQIDKKLDITPIKEMEDIEIETDEKTLFGKNKTEIVRQWTGNIILSENDYLKLNKEIKKGKK
TEGRLAAILETDVYQENKELKNELKD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 4085 | GenBank | NZ_CP116964 |
| Plasmid name | pDM86-optrA | Incompatibility group | - |
| Plasmid size | 54078 bp | Coordinate of oriT [Strand] | 36872..36912 [+] |
| Host baterium | Enterococcus faecalis strain DM86 |
Cargo genes
| Drug resistance gene | aac(6')-aph(2''), aph(2'')-Ia, erm(A), optrA, fexA, aadD, ant(9)-Ia |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |