Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103489 |
| Name | oriT_AGR_348|unnamed1 |
| Organism | Serratia grimesii strain AGR_348 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MT039164 (26044..26125 [-], 82 nt) |
| oriT length | 82 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 82 nt
>oriT_AGR_348|unnamed1
CGGGACCAGATGTGTTTTGTAGCACCGCCTGCACGCAGTCGGTTCAACAAAACGCTGGGTCAGGGCAGAGCCCCGACACCCC
CGGGACCAGATGTGTTTTGTAGCACCGCCTGCACGCAGTCGGTTCAACAAAACGCTGGGTCAGGGCAGAGCCCCGACACCCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file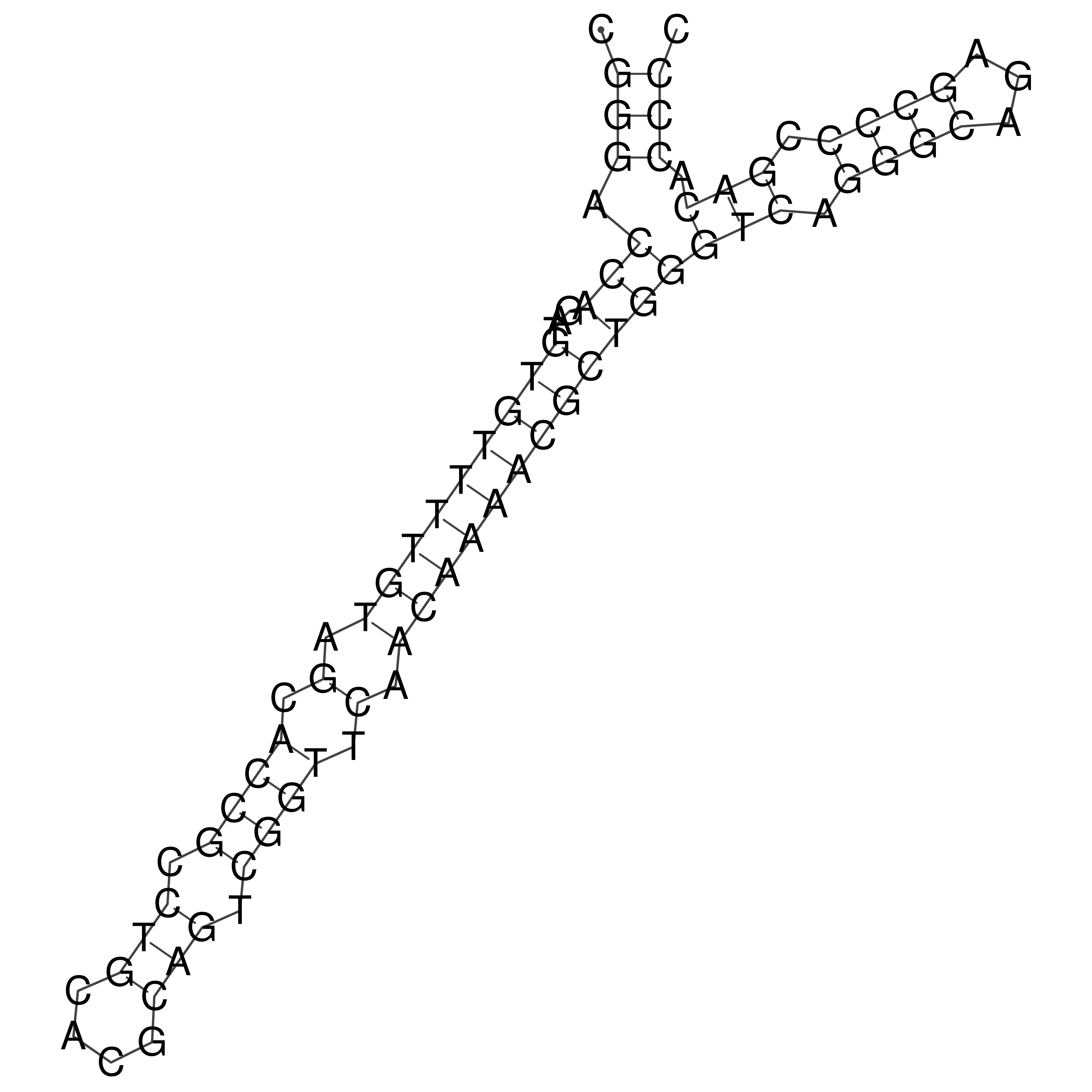
Relaxase
| ID | 2611 | GenBank | WP_241390007 |
| Name | Relaxase_MOO72_RS00115_AGR_348|unnamed1 |
UniProt ID | _ |
| Length | 651 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 651 a.a. Molecular weight: 73413.65 Da Isoelectric Point: 7.3733
>WP_241390007.1 TraI/MobA(P) family conjugative relaxase [Serratia grimesii]
MIIRIPEKRRDGKSSFLQLVAYTVVRDEDKPDTPLEPEHPGWRRPRSKDAIFNHLVDYISRNGDADLQQI
MHADPHGHTQVMFDGVMCETNCFNITTASVEMNAVALQNTRCKDPVLHYFLSWPETDNPSTEHIFDSVRH
SLSALGMSEHQYVAAIHTDTNNIHCHVAANRIHPETYKAADDSFTRIRLQQAARELELKYNWTPTNGFFA
VNEQGEIVRSKREKSLTPSGAKALEYYADVESLHTYAVTECGFKIDEAMADPNLTWKDVHRILVNAGLAL
KPKGKGLAIYSIDNPELPPVKASSVHPDLTLGCLEKDLGLFQPLENVGTYTKDHTTTATDAIVHEFRYEP
TLHARDLTARLERRLARADARADLKARYQAYKHTWKRPRLDASVVKRRYQNESKRFAWQKARARVAVGDP
LLRKLTYHIIEVERMKAMAALRLAVKEERAAFKADPANRRLSYREWVEQQALSHDQAAIAQLRAFAYRMK
RTQRTAPISTNSIVCAVADDTPAFALEGYATQVTRDGTVQYVRDGCVELQDKGERIEVGNHRVNEGEHIA
GGMALAENKSGEHLKFEGESAFVQQACSLVRWFNEGGDTPLPLSDPQQRILAGYDASSQAISTGEGQHKL
SAAEQPDVEQSKPRSGYRPQQ
MIIRIPEKRRDGKSSFLQLVAYTVVRDEDKPDTPLEPEHPGWRRPRSKDAIFNHLVDYISRNGDADLQQI
MHADPHGHTQVMFDGVMCETNCFNITTASVEMNAVALQNTRCKDPVLHYFLSWPETDNPSTEHIFDSVRH
SLSALGMSEHQYVAAIHTDTNNIHCHVAANRIHPETYKAADDSFTRIRLQQAARELELKYNWTPTNGFFA
VNEQGEIVRSKREKSLTPSGAKALEYYADVESLHTYAVTECGFKIDEAMADPNLTWKDVHRILVNAGLAL
KPKGKGLAIYSIDNPELPPVKASSVHPDLTLGCLEKDLGLFQPLENVGTYTKDHTTTATDAIVHEFRYEP
TLHARDLTARLERRLARADARADLKARYQAYKHTWKRPRLDASVVKRRYQNESKRFAWQKARARVAVGDP
LLRKLTYHIIEVERMKAMAALRLAVKEERAAFKADPANRRLSYREWVEQQALSHDQAAIAQLRAFAYRMK
RTQRTAPISTNSIVCAVADDTPAFALEGYATQVTRDGTVQYVRDGCVELQDKGERIEVGNHRVNEGEHIA
GGMALAENKSGEHLKFEGESAFVQQACSLVRWFNEGGDTPLPLSDPQQRILAGYDASSQAISTGEGQHKL
SAAEQPDVEQSKPRSGYRPQQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2412 | GenBank | WP_262190542 |
| Name | trbC_MOO72_RS00100_AGR_348|unnamed1 |
UniProt ID | _ |
| Length | 725 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 725 a.a. Molecular weight: 81720.52 Da Isoelectric Point: 8.5434
>WP_262190542.1 F-type conjugative transfer protein TrbC [Serratia grimesii]
MSRPVEVDPRRVRRPLGYTFINDALLSPIGVQLSLLAGLIVGFVMPATLLLTVPGLLLLVMLFADRPFRM
PLRMPTDIGGMDLTTEREMPKYRKGLGGFFRYVVRTRKYMPAAGVMCLGYARGKGLARELWLTLDDALRH
MLLLATTGSGKTEALLSVFLNSICWGRGICYSDGKGQNTLAFAMWSLARRFGREDDFYVLNFMTGGTDKL
LQLLLNDKKRQPSNTINLFGTANTTFIIQLMESMLPPAGSGDQGWQDKAKSMLSALVYAIYYKCKREKRR
ISQKVIQEYLPLRKLADLYQEAKRDGWHREAYNPLENYFNTLAGFRIELIKKPSEWEQGVYDQHGYLIQQ
FNRMLTMFNDLYGHIFSTDAGDIDIEDVLHNDRILCTTIPALELSKGEASNIGKLYISAIRMTMARDLGC
ELEGMMNDVLIVKKYSGKFPYPIAMDELGAYFGPGMDNLASQMRSLGYMLIVSAQDIQRFIAEHKGEYMT
VNANLLTKWFMTLQDEKDTFDLARITGGKGYYAELGSVEQAGGFITPNYEDAANNYIREKDRLDLGDLKD
MSPGEGMISFKSALVPSNAIYIPDDQKMTSSLPMRINRFIDVETPTEAELFTLNPSLKRKLPPTAQEIDG
ILQRLEQAMDTTTSAGMMDPVLKRVAAVALDLDNRADISYSPTQRGVLLFEAAREALHQNKRNWRQLPSP
PKPIRVSKEVAQSLANASTNQITFR
MSRPVEVDPRRVRRPLGYTFINDALLSPIGVQLSLLAGLIVGFVMPATLLLTVPGLLLLVMLFADRPFRM
PLRMPTDIGGMDLTTEREMPKYRKGLGGFFRYVVRTRKYMPAAGVMCLGYARGKGLARELWLTLDDALRH
MLLLATTGSGKTEALLSVFLNSICWGRGICYSDGKGQNTLAFAMWSLARRFGREDDFYVLNFMTGGTDKL
LQLLLNDKKRQPSNTINLFGTANTTFIIQLMESMLPPAGSGDQGWQDKAKSMLSALVYAIYYKCKREKRR
ISQKVIQEYLPLRKLADLYQEAKRDGWHREAYNPLENYFNTLAGFRIELIKKPSEWEQGVYDQHGYLIQQ
FNRMLTMFNDLYGHIFSTDAGDIDIEDVLHNDRILCTTIPALELSKGEASNIGKLYISAIRMTMARDLGC
ELEGMMNDVLIVKKYSGKFPYPIAMDELGAYFGPGMDNLASQMRSLGYMLIVSAQDIQRFIAEHKGEYMT
VNANLLTKWFMTLQDEKDTFDLARITGGKGYYAELGSVEQAGGFITPNYEDAANNYIREKDRLDLGDLKD
MSPGEGMISFKSALVPSNAIYIPDDQKMTSSLPMRINRFIDVETPTEAELFTLNPSLKRKLPPTAQEIDG
ILQRLEQAMDTTTSAGMMDPVLKRVAAVALDLDNRADISYSPTQRGVLLFEAAREALHQNKRNWRQLPSP
PKPIRVSKEVAQSLANASTNQITFR
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 783..20577
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MOO72_RS00005 (348p1_00001) | 1..783 | + | 783 | WP_336251214 | plasmid transfer ATPase TraJ | - |
| MOO72_RS00010 (348p1_00002) | 783..1049 | + | 267 | WP_241389991 | IcmT/TraK family protein | traK |
| MOO72_RS00015 (348p1_00004) | 1192..1530 | + | 339 | WP_241389992 | DUF5710 domain-containing protein | - |
| MOO72_RS00020 (348p1_00005) | 1488..4424 | + | 2937 | WP_241389993 | LPD7 domain-containing protein | - |
| MOO72_RS00025 (348p1_00006) | 4434..4925 | + | 492 | WP_241389994 | hypothetical protein | traL |
| MOO72_RS00030 (348p1_00007) | 4949..5692 | + | 744 | WP_241389995 | DotI/IcmL/TraM family protein | traM |
| MOO72_RS00035 (348p1_00009) | 5946..6701 | + | 756 | WP_241390046 | DotH/IcmK family type IV secretion protein | traN |
| MOO72_RS00040 (348p1_00010) | 6706..7938 | + | 1233 | WP_127147003 | conjugal transfer protein TraO | traO |
| MOO72_RS00045 (348p1_00011) | 7938..8681 | + | 744 | WP_241389996 | hypothetical protein | traP |
| MOO72_RS00050 (348p1_00012) | 8690..9220 | + | 531 | WP_010895872 | conjugal transfer protein TraQ | traQ |
| MOO72_RS00055 (348p1_00013) | 9236..9637 | + | 402 | WP_241389997 | DUF6750 family protein | traR |
| MOO72_RS00060 (348p1_00014) | 9722..10327 | + | 606 | WP_241389998 | hypothetical protein | traT |
| MOO72_RS00065 (348p1_00015) | 10330..13404 | + | 3075 | WP_241389999 | type IV secretory system conjugative DNA transfer family protein | traU |
| MOO72_RS00070 (348p1_00017) | 13545..14771 | + | 1227 | WP_241390000 | conjugal transfer protein TraW | traW |
| MOO72_RS00075 (348p1_00018) | 14764..15327 | + | 564 | WP_241390001 | conjugal transfer protein TraX | - |
| MOO72_RS00080 (348p1_00019) | 15260..17536 | + | 2277 | WP_336251215 | DotA/TraY family protein | - |
| MOO72_RS00085 (348p1_00020) | 17533..18111 | + | 579 | WP_241390002 | hypothetical protein | - |
| MOO72_RS00090 (348p1_00021) | 18291..19550 | + | 1260 | WP_241390003 | conjugal transfer protein TrbA | trbA |
| MOO72_RS00095 (348p1_00022) | 19564..20577 | + | 1014 | WP_241390004 | hypothetical protein | trbB |
| MOO72_RS00100 (348p1_00023) | 20615..22792 | + | 2178 | WP_262190542 | F-type conjugative transfer protein TrbC | - |
| MOO72_RS00105 (348p1_00024) | 22803..23339 | + | 537 | WP_241390006 | phospholipase D family protein | - |
| MOO72_RS00110 (348p1_00025) | 23349..23672 | + | 324 | WP_127147014 | hypothetical protein | - |
Host bacterium
| ID | 3932 | GenBank | NZ_MT039164 |
| Plasmid name | AGR_348|unnamed1 | Incompatibility group | - |
| Plasmid size | 149773 bp | Coordinate of oriT [Strand] | 26044..26125 [-] |
| Host baterium | Serratia grimesii strain AGR_348 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | pilW |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |