Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103465 |
| Name | oriT_AGR_145|unnamed1 |
| Organism | Serratia proteamaculans strain AGR_145 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MT039183 (22259..22341 [+], 83 nt) |
| oriT length | 83 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 83 nt
>oriT_AGR_145|unnamed1
ACGGGACCAGATGTGTTTTGTAGCACCGCCTGCACGCAGTCGGTTCAACAAAACGCTGGGTCAGGGCAGAGCCCCGACACCCC
ACGGGACCAGATGTGTTTTGTAGCACCGCCTGCACGCAGTCGGTTCAACAAAACGCTGGGTCAGGGCAGAGCCCCGACACCCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file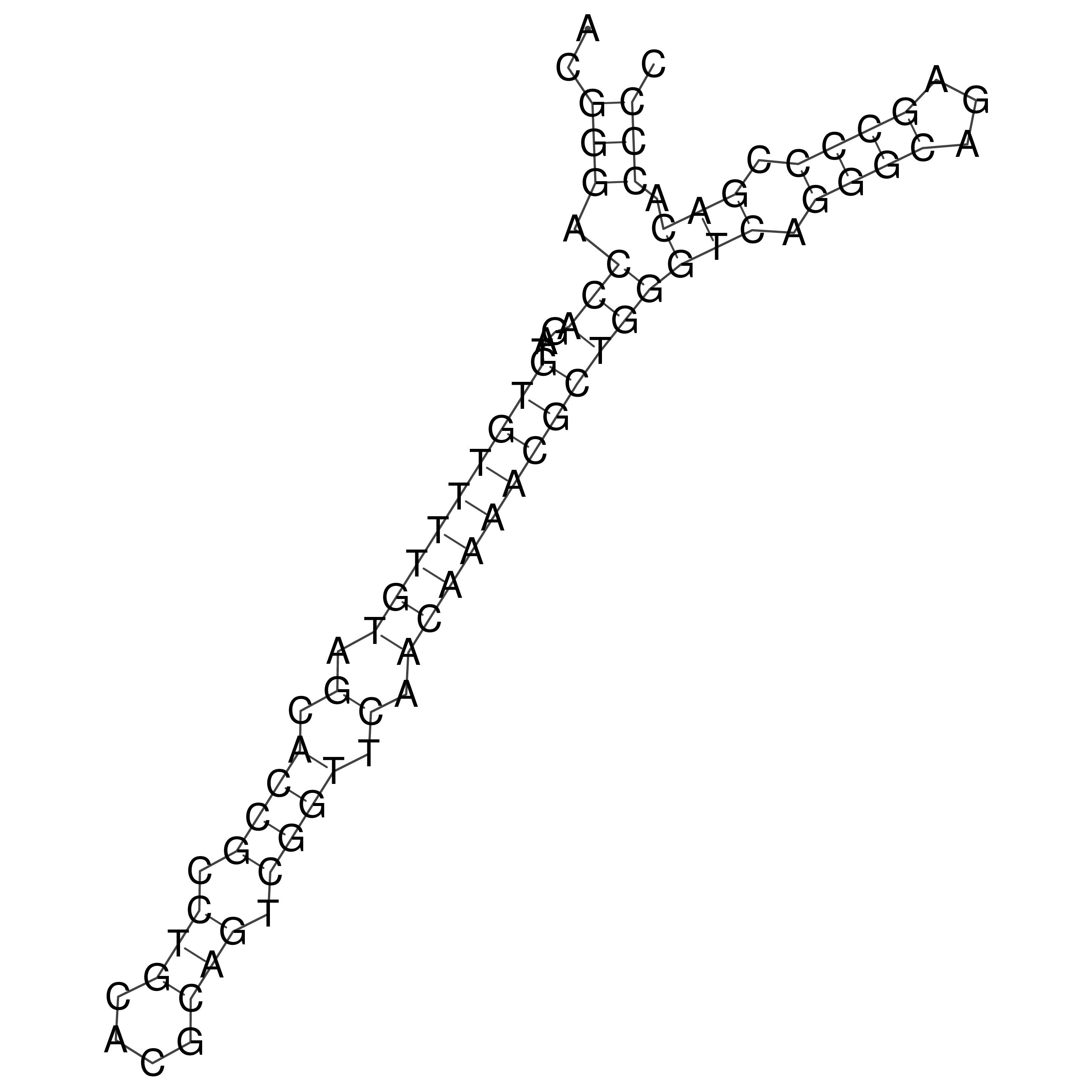
Relaxase
| ID | 2588 | GenBank | WP_010895859 |
| Name | Relaxase_MOP16_RS00140_AGR_145|unnamed1 |
UniProt ID | A7M7H3 |
| Length | 651 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 651 a.a. Molecular weight: 73395.58 Da Isoelectric Point: 7.4747
>WP_010895859.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Serratia]
MIIRIPEKRRDGKSSFLQLVAYTVVRDEDKPDSPLEPEHPGWRRPRSKDAIFNHLVDYISRNGDADLQQI
MHADPNGHTQVMFDGVMCETNCFNIATASVEMNAVALQNTRCKDPVLHYFLSWPETDNPSTEHIFDSVRH
SLSALGMSEHQYVAAIHTDTNNIHCHVAANRIHPETYKAADDSFTRIRLQQAARELELKYNWTPTNGFFA
VNEQGEIVRSKREKNLTPSGAKTLEYYADVESLHTYAVTECGFKIDEAMADPNLAWKDIHCILVNAGLAL
KPKGKGLAIYSIDNPELPPVKASSVHPDLTLNCLEKDLGQFQLLENAGTYTKEHTTTAADAIVHEFRYEP
TLHARDLTARLERRLARADARADLKARYQAYKNAWKRPKLDAGAVKRRYQNESKRFAWQKARARVAIGDP
LLRKLTYHIIEVERMKAMAALRLTVKDERAAFKADPANRRLSYREWVEQQALSHDQAAIAQLRAFAYRMK
RTQRTAPLSTNSIVCAVADDTPAFALEGYATRVTRDGTVQYVRDGRVQLQDKGERIDVGDHRVNEGEHIA
GGMALAENKSGEHLKFEGEAAFVQQACSMVPWFNEGGDAPLPLSDPQQRTMAGYESTYQSASTGEGQRKP
SAVEQPEVEQNKPRSGHRPQQ
MIIRIPEKRRDGKSSFLQLVAYTVVRDEDKPDSPLEPEHPGWRRPRSKDAIFNHLVDYISRNGDADLQQI
MHADPNGHTQVMFDGVMCETNCFNIATASVEMNAVALQNTRCKDPVLHYFLSWPETDNPSTEHIFDSVRH
SLSALGMSEHQYVAAIHTDTNNIHCHVAANRIHPETYKAADDSFTRIRLQQAARELELKYNWTPTNGFFA
VNEQGEIVRSKREKNLTPSGAKTLEYYADVESLHTYAVTECGFKIDEAMADPNLAWKDIHCILVNAGLAL
KPKGKGLAIYSIDNPELPPVKASSVHPDLTLNCLEKDLGQFQLLENAGTYTKEHTTTAADAIVHEFRYEP
TLHARDLTARLERRLARADARADLKARYQAYKNAWKRPKLDAGAVKRRYQNESKRFAWQKARARVAIGDP
LLRKLTYHIIEVERMKAMAALRLTVKDERAAFKADPANRRLSYREWVEQQALSHDQAAIAQLRAFAYRMK
RTQRTAPLSTNSIVCAVADDTPAFALEGYATRVTRDGTVQYVRDGRVQLQDKGERIDVGDHRVNEGEHIA
GGMALAENKSGEHLKFEGEAAFVQQACSMVPWFNEGGDAPLPLSDPQQRTMAGYESTYQSASTGEGQRKP
SAVEQPEVEQNKPRSGHRPQQ
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A7M7H3 |
T4CP
| ID | 2387 | GenBank | WP_234591458 |
| Name | trbC_MOP16_RS00155_AGR_145|unnamed1 |
UniProt ID | _ |
| Length | 725 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 725 a.a. Molecular weight: 81497.36 Da Isoelectric Point: 8.5425
>WP_234591458.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Serratia]
MSRPVEVDPRRVRRPLGYTFINDALLSPMGVQLSLLAGLIAGFVMPATLLLTVPGLLLLVMLFADRPFRM
PLRMPTDIGGMDLTTEREMPKYRKGLGGFFRYVVRTRKYMPAAGVMCLGYARGKGLARELWLTLDDALRH
MLLLATTGSGKTEALLSVFLNSICWGRGICYSDGKGQNTLAFAMWSLARRFGREDDFYVLNFMTGGTDKL
LQLLLNDKKRQPSNTINLFGTANTTFIIQLMESMLPPAGSGDQGWQDKAKSMLSALVYAIYYKCKREKRR
ISQKVIQEYLPLRKLADLYQEAKRDGWHREAYNPLENYFNTLAGFRIELISKPSEWEQGVYDQHGYLIQQ
FNRMLTMFNDLYGHIFSTDAGDIDIEDVLHNDRILCTTIPALELSKGEASNIGKLYISAIRMTMARDLGC
ELEGMMNDVLIVKKYSGKFPYPIAMDELGAYFGPGMDNLASQMRSLGYMLIVSAQDIQRFIAEHKGEYMT
VNANLLTKWFMTLQDEKDTFELARITGGKGYYAELGSVEQAGGFITPNYEDAANNYIREKDRLDLGDLKD
MSPGEGMISFKSALVPSNAIYIPDDQKMTSALPMRINRFIDVETPTEAELFALNPSLKRKLPPTAQEVDG
ILQRLDLAVDTTASVGIMDPVLKRVAAVALDLDNRADVSYSPAQRGVLLFEAAREALHKNKRKWRQLPTP
PKPIRVSKEVAQSLANAGTNEITFR
MSRPVEVDPRRVRRPLGYTFINDALLSPMGVQLSLLAGLIAGFVMPATLLLTVPGLLLLVMLFADRPFRM
PLRMPTDIGGMDLTTEREMPKYRKGLGGFFRYVVRTRKYMPAAGVMCLGYARGKGLARELWLTLDDALRH
MLLLATTGSGKTEALLSVFLNSICWGRGICYSDGKGQNTLAFAMWSLARRFGREDDFYVLNFMTGGTDKL
LQLLLNDKKRQPSNTINLFGTANTTFIIQLMESMLPPAGSGDQGWQDKAKSMLSALVYAIYYKCKREKRR
ISQKVIQEYLPLRKLADLYQEAKRDGWHREAYNPLENYFNTLAGFRIELISKPSEWEQGVYDQHGYLIQQ
FNRMLTMFNDLYGHIFSTDAGDIDIEDVLHNDRILCTTIPALELSKGEASNIGKLYISAIRMTMARDLGC
ELEGMMNDVLIVKKYSGKFPYPIAMDELGAYFGPGMDNLASQMRSLGYMLIVSAQDIQRFIAEHKGEYMT
VNANLLTKWFMTLQDEKDTFELARITGGKGYYAELGSVEQAGGFITPNYEDAANNYIREKDRLDLGDLKD
MSPGEGMISFKSALVPSNAIYIPDDQKMTSALPMRINRFIDVETPTEAELFALNPSLKRKLPPTAQEVDG
ILQRLDLAVDTTASVGIMDPVLKRVAAVALDLDNRADVSYSPAQRGVLLFEAAREALHKNKRKWRQLPTP
PKPIRVSKEVAQSLANAGTNEITFR
Protein domains
No domain identified.
Protein structure
No available structure.
| ID | 2388 | GenBank | WP_010895767 |
| Name | t4cp2_MOP16_RS00350_AGR_145|unnamed1 |
UniProt ID | _ |
| Length | 577 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 577 a.a. Molecular weight: 64155.03 Da Isoelectric Point: 5.2094
>WP_010895767.1 MULTISPECIES: ATP-binding protein [Serratia]
MPIFNFRDEDALGKVASVDTTNVIVDVENVEQLKRLQVNHLAVLQSSRPGQHLIGLITQVTRKRGIEDIA
NDGISEQTSELNLCKIALIGTMLDRDGERENVFRRTLESVPEIDANCFSLEGQNLTNFMRTLSSVSADGN
ALTLGKYTLDEHAIAYLNGNKFFQRHAFIGGSTGSGKSWTTAKIIEQMAGLSTANAIVFDLHGEYSPLVG
KGIQHFKVAGPADVEAQRTIDNGVLYLPYWLLSYEALVAMFVDRSDQNAPNQAMIMSREINQAKRKYLED
GDHKEILKHFTVDSPVPFDLDVLMGRLNEINVEMVPGASQGKEKQGDFFGKLARMISRLENKISDRRLGF
IFSGGGDVLNFSWLEKFTTAVLGSSEENGNAGIKIINFSEVPSDVLPLIVSLVARVTFSVQQWTPSELRH
PIALLCDEAHLYMPQRNMAESADDISLDIFERIAKEGRKYGVSLIVISQRPSEVNKTMLSQCSNFVSMRL
TNAEDQGVIKRLLPDSLGGFSDILPTLDTGEALVVGDASLLPSRIRIDEPINKPNSGTVNFWDEWQKPVK
DKRLLIAVENWRKQNIQ
MPIFNFRDEDALGKVASVDTTNVIVDVENVEQLKRLQVNHLAVLQSSRPGQHLIGLITQVTRKRGIEDIA
NDGISEQTSELNLCKIALIGTMLDRDGERENVFRRTLESVPEIDANCFSLEGQNLTNFMRTLSSVSADGN
ALTLGKYTLDEHAIAYLNGNKFFQRHAFIGGSTGSGKSWTTAKIIEQMAGLSTANAIVFDLHGEYSPLVG
KGIQHFKVAGPADVEAQRTIDNGVLYLPYWLLSYEALVAMFVDRSDQNAPNQAMIMSREINQAKRKYLED
GDHKEILKHFTVDSPVPFDLDVLMGRLNEINVEMVPGASQGKEKQGDFFGKLARMISRLENKISDRRLGF
IFSGGGDVLNFSWLEKFTTAVLGSSEENGNAGIKIINFSEVPSDVLPLIVSLVARVTFSVQQWTPSELRH
PIALLCDEAHLYMPQRNMAESADDISLDIFERIAKEGRKYGVSLIVISQRPSEVNKTMLSQCSNFVSMRL
TNAEDQGVIKRLLPDSLGGFSDILPTLDTGEALVVGDASLLPSRIRIDEPINKPNSGTVNFWDEWQKPVK
DKRLLIAVENWRKQNIQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 27934..56225
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MOP16_RS00145 (145p_00037) | 24709..25032 | - | 324 | WP_010895860 | hypothetical protein | - |
| MOP16_RS00150 (145p_00038) | 25042..25578 | - | 537 | WP_234591459 | phospholipase D family protein | - |
| MOP16_RS00155 (145p_00039) | 25719..27896 | - | 2178 | WP_234591458 | F-type conjugative transfer protein TrbC | - |
| MOP16_RS00160 (145p_00040) | 27934..28932 | - | 999 | WP_010895863 | hypothetical protein | trbB |
| MOP16_RS00165 (145p_00041) | 28945..30255 | - | 1311 | WP_241389809 | conjugal transfer protein TrbA | trbA |
| MOP16_RS00170 (145p_00043) | 30393..30893 | - | 501 | WP_241389810 | hypothetical protein | - |
| MOP16_RS00175 (145p_00044) | 30971..33265 | - | 2295 | WP_241389811 | DotA/TraY family protein | traY |
| MOP16_RS00180 (145p_00045) | 33180..33743 | - | 564 | WP_010895867 | conjugal transfer protein TraX | - |
| MOP16_RS00185 (145p_00046) | 33736..34962 | - | 1227 | WP_010895868 | conjugal transfer protein TraW | traW |
| MOP16_RS00190 (145p_00048) | 35103..38177 | - | 3075 | WP_241389812 | type IV secretory system conjugative DNA transfer family protein | traU |
| MOP16_RS00195 (145p_00049) | 38180..38785 | - | 606 | WP_010895870 | TraT | traT |
| MOP16_RS00200 (145p_00050) | 38871..39272 | - | 402 | WP_010895871 | DUF6750 family protein | traR |
| MOP16_RS00205 (145p_00051) | 39288..39818 | - | 531 | WP_010895872 | conjugal transfer protein TraQ | traQ |
| MOP16_RS00210 (145p_00052) | 39827..40570 | - | 744 | WP_241389813 | hypothetical protein | traP |
| MOP16_RS00215 (145p_00053) | 40570..41802 | - | 1233 | WP_010895874 | conjugal transfer protein TraO | traO |
| MOP16_RS00220 (145p_00054) | 41807..42814 | - | 1008 | WP_241389814 | DotH/IcmK family type IV secretion protein | traN |
| MOP16_RS00225 (145p_00055) | 42817..43491 | - | 675 | WP_241389815 | DotI/IcmL/TraM family protein | traM |
| MOP16_RS00230 (145p_00056) | 43582..44073 | - | 492 | WP_010895743 | hypothetical protein | traL |
| MOP16_RS00235 (145p_00057) | 44083..47319 | - | 3237 | WP_010895744 | LPD7 domain-containing protein | - |
| MOP16_RS00240 (145p_00059) | 47462..47728 | - | 267 | WP_010895745 | IcmT/TraK family protein | traK |
| MOP16_RS00245 (145p_00060) | 47728..48885 | - | 1158 | WP_010895746 | plasmid transfer ATPase TraJ | virB11 |
| MOP16_RS00250 (145p_00062) | 49075..49863 | - | 789 | WP_010895747 | type IV secretory system conjugative DNA transfer family protein | traI |
| MOP16_RS00255 (145p_00063) | 49860..50318 | - | 459 | WP_010895748 | DotD/TraH family lipoprotein | - |
| MOP16_RS00260 (145p_00064) | 50356..51789 | - | 1434 | WP_010895749 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| MOP16_RS00265 (145p_00065) | 51786..52454 | - | 669 | WP_241389817 | prepilin peptidase | - |
| MOP16_RS00270 (145p_00066) | 52454..52939 | - | 486 | WP_010895751 | lytic transglycosylase domain-containing protein | virB1 |
| MOP16_RS00275 (145p_00067) | 52994..53587 | - | 594 | WP_010895752 | type 4 pilus major pilin | - |
| MOP16_RS00280 (145p_00068) | 53623..54735 | - | 1113 | WP_241389818 | type II secretion system F family protein | - |
| MOP16_RS00285 (145p_00069) | 54735..56225 | - | 1491 | WP_010895754 | ATPase, T2SS/T4P/T4SS family | virB11 |
| MOP16_RS00290 (145p_00070) | 56231..56815 | - | 585 | WP_010895755 | type IV pilus biogenesis protein PilP | - |
| MOP16_RS00295 (145p_00071) | 56802..58112 | - | 1311 | WP_010895756 | type 4b pilus protein PilO2 | - |
| MOP16_RS00300 (145p_00072) | 58116..59750 | - | 1635 | WP_241390125 | PilN family type IVB pilus formation outer membrane protein | - |
| MOP16_RS00305 (145p_00073) | 59775..60200 | - | 426 | WP_010895758 | type IV pilus biogenesis protein PilM | - |
| MOP16_RS00310 (145p_00074) | 60200..61213 | - | 1014 | WP_241389819 | toxin co-regulated pilus biosynthesis Q family protein | - |
Host bacterium
| ID | 3908 | GenBank | NZ_MT039183 |
| Plasmid name | AGR_145|unnamed1 | Incompatibility group | - |
| Plasmid size | 151370 bp | Coordinate of oriT [Strand] | 22259..22341 [+] |
| Host baterium | Serratia proteamaculans strain AGR_145 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |