Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103429 |
| Name | oriT_pIB2020_L |
| Organism | Enterobacter kobei strain IB2020 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP059484 (132113..132197 [-], 85 nt) |
| oriT length | 85 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 85 nt
>oriT_pIB2020_L
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGTGTTTGTAATATCGTGCGTGCGCGGTATTACAAACACACATCCTGTCCCGTCTTT
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGTGTTTGTAATATCGTGCGTGCGCGGTATTACAAACACACATCCTGTCCCGTCTTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file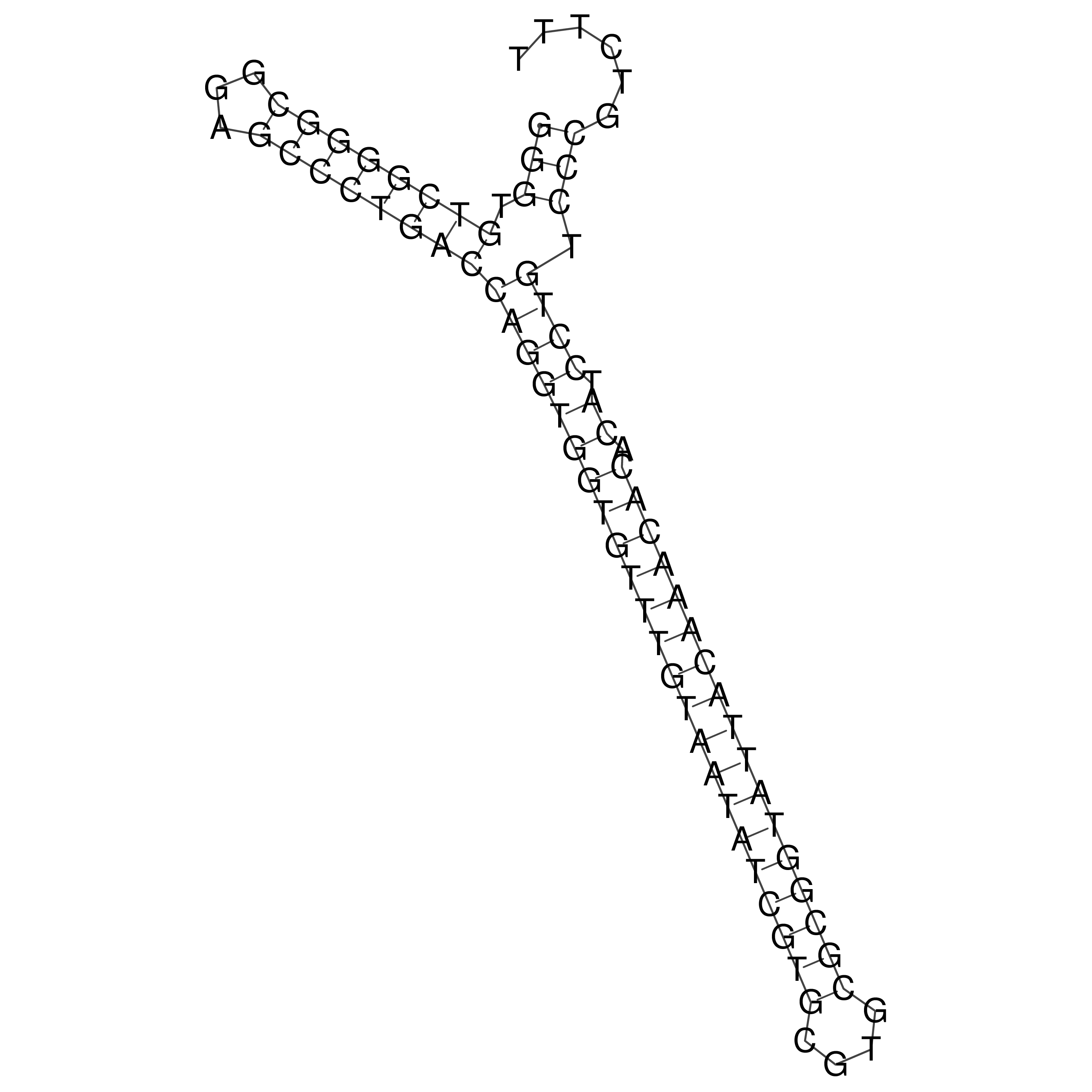
Relaxase
| ID | 2556 | GenBank | WP_048242245 |
| Name | Relaxase_H1R18_RS26380_pIB2020_L |
UniProt ID | A0AA36ETS3 |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 103346.66 Da Isoelectric Point: 8.3785
>WP_048242245.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterobacteriaceae]
MIAVIPPKRKDGKSSFVDLVSYVSVRDEERDDDLMEAVKASGMKPEMPHRSRFSRLVDYATKLRDESFVS
LVDVMPDGGEWVNFYGVTCFHNCTSIETAAEEMEFIAQKARYAHDNSDPVFHYLLSWQSHESPRPEQVYD
SVRHSLKRLGMSDHQFVAAIHTDTDNLHVHVAVNRVHPETGYLNWLSYSQEKLHKACRELELKHGFSPDN
GCYVIAPDNRIIRRTSVERDRQSAWTRGRNQSLKEYIADTTIAGLRESPVTDWSSLHQRFAKEGLFLSAD
NGELKVKDGWNSERPGVTLSTFGHAFDTDKLIRKMGEFIPPAQDIFAQVPAVGRYEPDKVIVPSRPERMT
EKESLSDYAIARLRAPLIALDQDPSQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDPTRTPVRAERVWP
ALTKAILDSYKGGWQPVPKDIFRQVTPTEQFTGGGLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAISHCRQEIETLIGTGQFTWERCHEQFARQGLLLMREHQGLVVMDAYNHEQTPVKASHVHPDLTLAR
AEPHAGAFRPVPADIFKRVPPESRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WRKPDLRGRERYQDIHEMCRRRKAYIRAQYRDPLIRKLHYHIAEVQRMQALIKLKETMKDERLQLIGEGK
WYPPSYRQWVEVEALKGDKAAISQLRGWDYRDRRGDAVKVTTDKRCVVICEPGGTPVYAGVPGLKAALQK
NGRVQFRDEETGEHICTDYGDRVIFGNSRDYDTLKRNMVKVSPILFSRSPDVAMSPQGDNEQFNQAFAKM
VAWHNVNRQDGDEYRISRADIDRLRVESEQQFMGMSGQGNDSRYDRDEPKQTWKPPSPL
MIAVIPPKRKDGKSSFVDLVSYVSVRDEERDDDLMEAVKASGMKPEMPHRSRFSRLVDYATKLRDESFVS
LVDVMPDGGEWVNFYGVTCFHNCTSIETAAEEMEFIAQKARYAHDNSDPVFHYLLSWQSHESPRPEQVYD
SVRHSLKRLGMSDHQFVAAIHTDTDNLHVHVAVNRVHPETGYLNWLSYSQEKLHKACRELELKHGFSPDN
GCYVIAPDNRIIRRTSVERDRQSAWTRGRNQSLKEYIADTTIAGLRESPVTDWSSLHQRFAKEGLFLSAD
NGELKVKDGWNSERPGVTLSTFGHAFDTDKLIRKMGEFIPPAQDIFAQVPAVGRYEPDKVIVPSRPERMT
EKESLSDYAIARLRAPLIALDQDPSQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDPTRTPVRAERVWP
ALTKAILDSYKGGWQPVPKDIFRQVTPTEQFTGGGLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAISHCRQEIETLIGTGQFTWERCHEQFARQGLLLMREHQGLVVMDAYNHEQTPVKASHVHPDLTLAR
AEPHAGAFRPVPADIFKRVPPESRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WRKPDLRGRERYQDIHEMCRRRKAYIRAQYRDPLIRKLHYHIAEVQRMQALIKLKETMKDERLQLIGEGK
WYPPSYRQWVEVEALKGDKAAISQLRGWDYRDRRGDAVKVTTDKRCVVICEPGGTPVYAGVPGLKAALQK
NGRVQFRDEETGEHICTDYGDRVIFGNSRDYDTLKRNMVKVSPILFSRSPDVAMSPQGDNEQFNQAFAKM
VAWHNVNRQDGDEYRISRADIDRLRVESEQQFMGMSGQGNDSRYDRDEPKQTWKPPSPL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2346 | GenBank | WP_048242233 |
| Name | trbC_H1R18_RS26425_pIB2020_L |
UniProt ID | _ |
| Length | 762 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 762 a.a. Molecular weight: 86870.32 Da Isoelectric Point: 6.8284
>WP_048242233.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MQNTPVNQAMINRNAWGSPLIELLRDHLLYAVIMAGSMVAGFIWPLSIPLCLLLAIVASISFSTHRWRMP
MRIPLYLHRVDPSEDRKVRRSLFKAFPSLFQYETLQESTGRGIFYLGYQRLRDTGRELWLSLDDLTRHVM
FFAGTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTVWYLALRFGREDDVEFINFMTGGMSRSEI
IHKGDKSRPQSNTFNPFGLSTEAFIAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKYWCVRENKTMSL
QLLREFMPLEKLAELYCRAVDDQWPEEAVSPLHNYLVDVPGFDMALVRTPSAWTEEPRKQHSYLTGQFLE
TFSTFTETFGDIFAEDAGDIDIRDSIHSDRILLTLIPAMNTSQHTTSALGRMFVTQQSMILARDLGYRLE
GLDTEALEITKYKGQFPYMNYLDEVGAFYTERIAVQATQVRSLEFALVMMAQDQERIETQTSAANVATLM
QNAGTKIAGKIVSEDKTAQTISNAAGKEARASMLNLQRRDGVIGTSWIDGDHINIQMENKINVQDLMKLQ
PGEHYTVFQGDPVPGASFYISPEEKSCNIPLVINRYITVNPPKLERLRRLVPRTAQRRLPTPENVSRIIG
VLTEKTSRRRKKTDTMPWKVIDTFQQRLANRQEAHNLLTDFDMDFAGREKNLWEEALDTIRTTTMAERTI
RYVMLNRPESSAPSREDDAPAPNALLKNLTLPPVTHVPVVRKRNDEPPVQAPPPLNQMEWPE
MQNTPVNQAMINRNAWGSPLIELLRDHLLYAVIMAGSMVAGFIWPLSIPLCLLLAIVASISFSTHRWRMP
MRIPLYLHRVDPSEDRKVRRSLFKAFPSLFQYETLQESTGRGIFYLGYQRLRDTGRELWLSLDDLTRHVM
FFAGTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTVWYLALRFGREDDVEFINFMTGGMSRSEI
IHKGDKSRPQSNTFNPFGLSTEAFIAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKYWCVRENKTMSL
QLLREFMPLEKLAELYCRAVDDQWPEEAVSPLHNYLVDVPGFDMALVRTPSAWTEEPRKQHSYLTGQFLE
TFSTFTETFGDIFAEDAGDIDIRDSIHSDRILLTLIPAMNTSQHTTSALGRMFVTQQSMILARDLGYRLE
GLDTEALEITKYKGQFPYMNYLDEVGAFYTERIAVQATQVRSLEFALVMMAQDQERIETQTSAANVATLM
QNAGTKIAGKIVSEDKTAQTISNAAGKEARASMLNLQRRDGVIGTSWIDGDHINIQMENKINVQDLMKLQ
PGEHYTVFQGDPVPGASFYISPEEKSCNIPLVINRYITVNPPKLERLRRLVPRTAQRRLPTPENVSRIIG
VLTEKTSRRRKKTDTMPWKVIDTFQQRLANRQEAHNLLTDFDMDFAGREKNLWEEALDTIRTTTMAERTI
RYVMLNRPESSAPSREDDAPAPNALLKNLTLPPVTHVPVVRKRNDEPPVQAPPPLNQMEWPE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3584..35434
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| H1R18_RS25620 (H1R18_25605) | 421..1242 | - | 822 | Protein_1 | DotA/TraY family protein | - |
| H1R18_RS25625 (H1R18_25610) | 1350..1511 | - | 162 | Protein_2 | transposase | - |
| H1R18_RS25630 (H1R18_25615) | 1527..2417 | - | 891 | WP_048242223 | Rpn family recombination-promoting nuclease/putative transposase | - |
| H1R18_RS25635 (H1R18_25620) | 2485..2964 | - | 480 | WP_048242221 | ATP-binding protein | - |
| H1R18_RS25640 (H1R18_25625) | 3048..3587 | - | 540 | WP_048242219 | conjugal transfer protein TraX | - |
| H1R18_RS25645 (H1R18_25630) | 3584..4786 | - | 1203 | WP_048242217 | conjugal transfer protein TraW | traW |
| H1R18_RS25650 (H1R18_25635) | 4801..5373 | - | 573 | WP_053071110 | hypothetical protein | traV |
| H1R18_RS25655 (H1R18_25640) | 5370..8423 | - | 3054 | WP_048242215 | conjugal transfer protein | traU |
| H1R18_RS25660 (H1R18_25645) | 8592..9410 | - | 819 | WP_048242213 | hypothetical protein | traT |
| H1R18_RS25665 (H1R18_25650) | 9397..9591 | - | 195 | WP_048242212 | hypothetical protein | - |
| H1R18_RS25670 (H1R18_25655) | 9640..10044 | - | 405 | WP_048242210 | DUF6750 family protein | traR |
| H1R18_RS25675 (H1R18_25660) | 10078..10605 | - | 528 | WP_048242208 | conjugal transfer protein TraQ | traQ |
| H1R18_RS25680 (H1R18_25665) | 10605..11369 | - | 765 | WP_048242206 | conjugal transfer protein TraP | traP |
| H1R18_RS25685 (H1R18_25670) | 11366..12673 | - | 1308 | WP_048242204 | conjugal transfer protein TraO | traO |
| H1R18_RS25690 (H1R18_25675) | 12676..13677 | - | 1002 | WP_048242202 | DotH/IcmK family type IV secretion protein | traN |
| H1R18_RS25695 (H1R18_25680) | 13688..14386 | - | 699 | WP_182115633 | DotI/IcmL family type IV secretion protein | traM |
| H1R18_RS25700 (H1R18_25685) | 14392..14745 | - | 354 | WP_048242199 | hypothetical protein | traL |
| H1R18_RS26905 (H1R18_25690) | 14763..19328 | - | 4566 | WP_313860150 | LPD7 domain-containing protein | - |
| H1R18_RS25710 (H1R18_25695) | 19513..20038 | - | 526 | Protein_19 | macro domain-containing protein | - |
| H1R18_RS25715 (H1R18_25700) | 20042..20488 | - | 447 | WP_048242192 | hypothetical protein | - |
| H1R18_RS25720 (H1R18_25705) | 20523..20810 | - | 288 | WP_048242190 | IcmT/TraK family protein | traK |
| H1R18_RS25725 (H1R18_25710) | 20807..21958 | - | 1152 | WP_152400182 | plasmid transfer ATPase TraJ | virB11 |
| H1R18_RS25730 (H1R18_25715) | 21942..22787 | - | 846 | WP_152400116 | type IV secretory system conjugative DNA transfer family protein | traI |
| H1R18_RS25735 (H1R18_25720) | 22784..23251 | - | 468 | WP_152400118 | DotD/TraH family lipoprotein | - |
| H1R18_RS25740 (H1R18_25725) | 23456..23713 | - | 258 | WP_152400120 | hypothetical protein | - |
| H1R18_RS25745 (H1R18_25730) | 23710..24357 | - | 648 | WP_152400123 | hypothetical protein | - |
| H1R18_RS25750 (H1R18_25735) | 24412..25602 | - | 1191 | WP_152400125 | conjugal transfer protein TraF | - |
| H1R18_RS25755 (H1R18_25740) | 25701..26549 | - | 849 | WP_152400127 | hypothetical protein | traE |
| H1R18_RS25760 (H1R18_25745) | 26630..27178 | - | 549 | WP_152400129 | hypothetical protein | - |
| H1R18_RS25765 (H1R18_25750) | 27204..28364 | - | 1161 | WP_152400131 | site-specific integrase | - |
| H1R18_RS25770 (H1R18_25755) | 29538..30911 | - | 1374 | WP_182115634 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| H1R18_RS25775 (H1R18_25760) | 30908..31573 | - | 666 | WP_152400133 | prepilin peptidase | - |
| H1R18_RS25780 (H1R18_25765) | 31577..32107 | - | 531 | WP_110820829 | lytic transglycosylase domain-containing protein | virB1 |
| H1R18_RS25785 (H1R18_25770) | 32117..32719 | - | 603 | WP_152400135 | type 4 pilus major pilin | - |
| H1R18_RS25790 (H1R18_25775) | 32741..33838 | - | 1098 | WP_152400137 | type II secretion system F family protein | - |
| H1R18_RS25795 (H1R18_25780) | 33869..35434 | - | 1566 | WP_152400139 | ATPase, T2SS/T4P/T4SS family | virB11 |
| H1R18_RS25800 (H1R18_25785) | 35442..35936 | - | 495 | WP_152400141 | type IV pilus biogenesis protein PilP | - |
| H1R18_RS25805 (H1R18_25790) | 35920..37221 | - | 1302 | WP_152400143 | type 4b pilus protein PilO2 | - |
| H1R18_RS25810 (H1R18_25795) | 37233..38918 | - | 1686 | WP_152400145 | PilN family type IVB pilus formation outer membrane protein | - |
| H1R18_RS25815 (H1R18_25800) | 38932..39369 | - | 438 | WP_152400147 | type IV pilus biogenesis protein PilM | - |
Host bacterium
| ID | 3872 | GenBank | NZ_CP059484 |
| Plasmid name | pIB2020_L | Incompatibility group | - |
| Plasmid size | 150133 bp | Coordinate of oriT [Strand] | 132113..132197 [-] |
| Host baterium | Enterobacter kobei strain IB2020 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIF6 |