Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103323 |
| Name | oriT_pVb1636 |
| Organism | Vibrio alginolyticus strain Vb1636 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MH548371 (47591..47696 [+], 106 nt) |
| oriT length | 106 nt |
| IRs (inverted repeats) | 66..73, 76..83 (CCACCAAC..GTTGGTGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 106 nt
AGCCTCGCAGAGCAGGATTCCCGTTGAGCCCGAAGGCGCGAATAAGGGTTAGAGTGAAGTAGCATCCACCAACTTGTTGGTGGGGCTACTTCACCTATCCTGCCCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file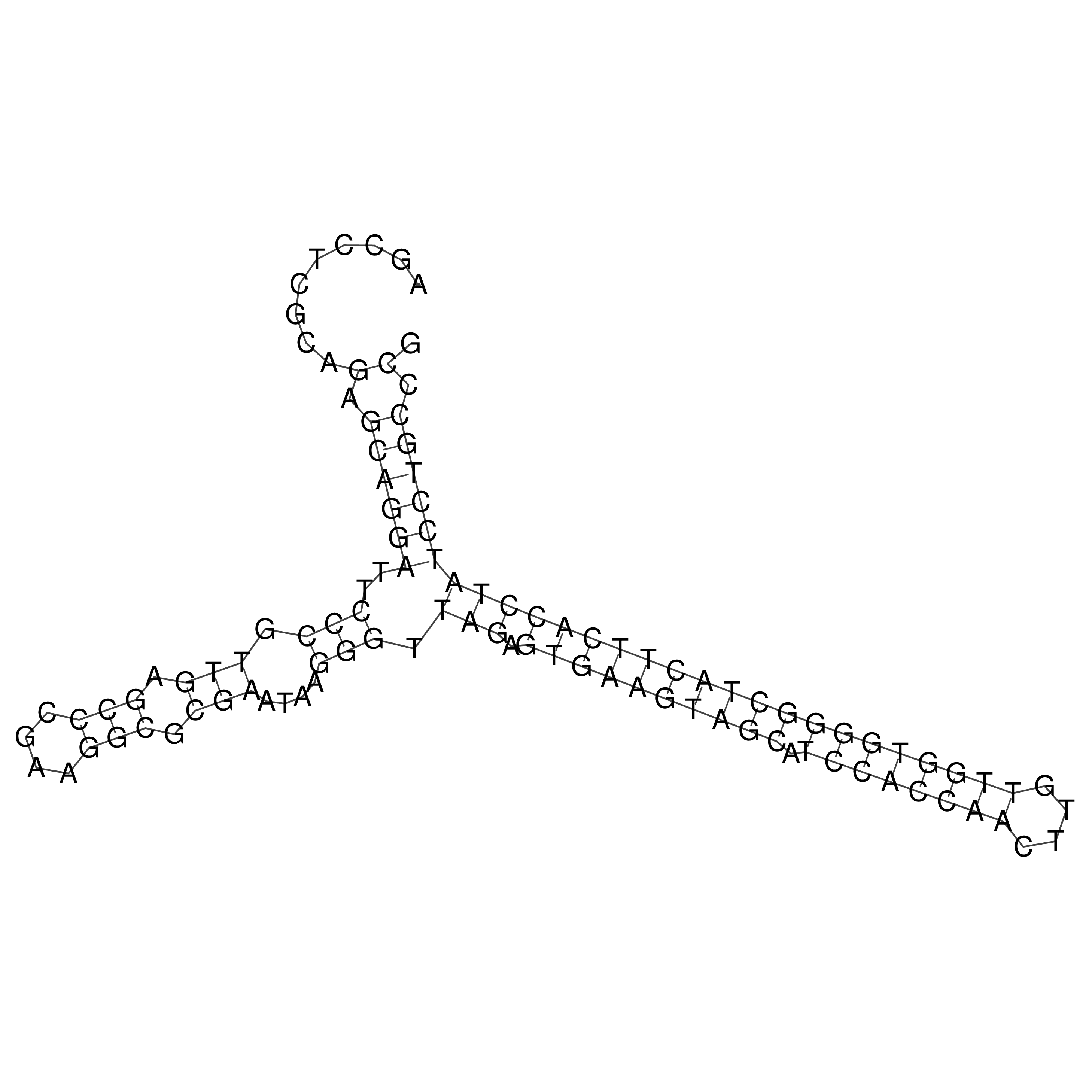
Relaxase
| ID | 2481 | GenBank | WP_169629002 |
| Name | Relaxase_HTT42_RS00240_pVb1636 |
UniProt ID | _ |
| Length | 798 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 798 a.a. Molecular weight: 88962.77 Da Isoelectric Point: 10.7371
MIIKHVPMRSAKKSDFSGLVKYITDEQSKTERLGLVNVTNCHANTMQAVIGEVLATQHTNTRATGDKTYH
LIVSFRPGEKPDSDVLKAIEERVCSGLGFSEHQRVSAVHHDTDNVHIHIAINKIHPTNKTMHEPFQAYRK
FGSLSDILENEYGLEKDNHQGFKTVSEGRASDMEKHAGIESLVSWVKRECLEEIRAAENWQQLHQVLSEN
SLEIRKQGNGFVVESADGTQVKASTLARDLSKQQLEKKLGEFEQINNPVPPQDKRRRYEKRPTKFKVNTT
ELYAKYQEEQKQLTAARKIEWINAKGRKDSRIEAAKRKNRLRRAAIKLMGESRLNKKILYSQAHKALKAD
IQAINKANKLERQALYEKYKRRAWADWLKQESLKGNKTALEALRARDKAKALKGDVISGKQKKSSPVNLE
GLNKPNVAPIGKKPPAFAKNRLRSMSSLGAVHFEHKPGYAPVMDNITKKGTIIYRAGKSAIRDDGEKLQI
SREATNEALKNALLLAKERYGSRITVNGSAQFKAQVIFTAATLNLPITFADAGMERRRQDLLNQENNHDR
REQTTGRTERGRSDRPSTSRLGRGITEHTDSGTRTTIANDDRSNTNGADSAPSTSNSVGGANAGNDGGNV
LRKPKPNVTGIGRNPPSIAKNRLRTLSSLSVVRFARRSEMLLPSDVSGQLEHQGTKPDNQLRWGVSGGGR
LNPGEVAVNKYLSERESKRAKGFDIPKHSRYNNQPGAFTYGGIRNVEGQALALLNRGDEVLVLPIDKATA
QRMKRVRIGEAVTVTPQGSLNKSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1066 | GenBank | WP_012774829 |
| Name | WP_012774829_pVb1636 |
UniProt ID | A0A510BQP9 |
| Length | 123 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 123 a.a. Molecular weight: 13720.78 Da Isoelectric Point: 7.1664
MTEEKKVTRKNSTPIKVYCLPDEKELIEAQAARAGMSAARYLREVGQGYHIQGVVDYDQVHELAKINGDL
GRLGGLLKLWLTNDVRTAQFGESTIRAVLSKIEATQEEMGAVMTKIVMPRSEQ
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A510BQP9 |
T4CP
| ID | 2268 | GenBank | WP_169629006 |
| Name | t4cp2_HTT42_RS00040_pVb1636 |
UniProt ID | _ |
| Length | 845 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 845 a.a. Molecular weight: 94343.99 Da Isoelectric Point: 6.2987
MIQSISFAIAGFGALLLLILFSRVRAVDAELKLKKHRSKDAGLADLLNYAAVVDDGVVVCKNGAFMASWL
YKGEDNASSTDAQREMVSFRINQALSGLGSGWMVHVDAVRRPSPKYSDRNLSEFPDKISAAIDEERRQLF
EGLGTMYEGYFVLTLTWFPPVLAQRKFVELMFDDDAEIVGKKEQTKGLINQFKREITAIENRLSSAVNLS
RLRGNKITNEDGKTITHDDFLGWLQFCITGLNHPVTLPSNPMYLDAYLGGQEMWGGVVPKVGRKFVQVVA
IEGFPMESYPGVLSALAELPVEYRWSSRFIFMDQHESLRHLDKFRKKWRQKIRGFFDQVFNTNNGTVDQD
ALSMVQDAEAAIAEISSGVVAAGYYTSVVVLMDENRDRLEQSARQVEKSVNRLGFAARTETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSTVWTGSNKAPCPMYPSLAPALMNCVTSGSTPFRLNLHVRDLGH
TFMFGPTGAGKSTHLALIAAQLRRYKGMSIYAFDKGMSMYPLTKAVNGLHFSVGADDDRLAFCPLQFLDS
KSDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIQSMYESGARTLSEFSLTIQDENIREAMRQYTVDGAM
GHLLDSEQDGLSLTDFTTFEIEELMNLGEKFALPVLLYLFRRIECSLKGQPSVIILDEAWLMLGHPAFRE
KIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMGLNSRQID
ILATAVPKRQYYYVSENGRRLYDLALGPLALSFVGASDKESVAIIKQLEAKHGEKWVDEWLASRGININN
YLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2269 | GenBank | WP_012774826 |
| Name | t4cp2_HTT42_RS00235_pVb1636 |
UniProt ID | _ |
| Length | 634 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 634 a.a. Molecular weight: 69997.69 Da Isoelectric Point: 6.8647
MNPKINNAVGPQVRAKKTSKNRTLPALGAVSIAVGLQSATQYFAHSFNYQESLGANINNVYAPWKILNWA
SDWYSNYPNQFMEAASVGMVVTSVGLLGVTITKVVTNNSSKANEYLHGSARWAGKKDIQSAGLLPRNRNL
LEVVTGKDAPSSTGVYVGGWEDEEGNFYYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWAQSAVITDLK
GELWALTAGWRQQYANNKVLRFEPASNYGGIHWNPLDEIRIGTEYEVGDVQNLATLIVDPDGKGMESHWQ
KTAFALLVGVILHALYKAKNEGTPATLPAVDAMLADPERDTGELWMEMATYGHVDGQNHPAVGSAARDMM
DRPEEEGGSVLSTAKSYLALYRDPVVARNVSESEFKIRDLMNHDNPVSLYIVTQPNDKARLRPLVRILVN
MIIRLLADEMTFEKGRPVAHYKHRLLAMLDEFPSLGKLEILQESLAFVAGYGIKCYLICQDLNQLKSRET
GYGPDETITSNCHVQNAYPPNRIETAEHLSRLTGQTTVVKEQITTSGRRTSALLGQVSRTIQEVQRPLLT
PDECLRMPGPVKNAKGDIDTAGDMVVYVAGYPAVYGKQPLYFKDPIFLARAQIEAPKVCDKLRVVEQVEE
GIQI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3585..14217
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTT42_RS00005 | 1..765 | + | 765 | WP_012774841 | replication initiation protein | - |
| HTT42_RS00010 | 1663..1887 | - | 225 | WP_172692984 | hypothetical protein | - |
| HTT42_RS00015 | 1874..2290 | - | 417 | WP_012774843 | hypothetical protein | - |
| HTT42_RS00020 | 2934..3311 | + | 378 | WP_012774845 | transcriptional regulator | - |
| HTT42_RS00025 | 3585..4574 | + | 990 | WP_012774846 | P-type conjugative transfer ATPase TrbB | virB11 |
| HTT42_RS00030 | 4586..5011 | + | 426 | WP_012774847 | conjugal transfer system pilin TrbC | virB2 |
| HTT42_RS00035 | 5014..5325 | + | 312 | WP_012774848 | conjugal transfer protein TrbD | virB3 |
| HTT42_RS00040 | 5325..7862 | + | 2538 | WP_169629006 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| HTT42_RS00045 | 7862..8638 | + | 777 | WP_012774850 | conjugal transfer protein TrbF | virB8 |
| HTT42_RS00050 | 8656..9561 | + | 906 | WP_169628991 | P-type conjugative transfer protein TrbG | virB9 |
| HTT42_RS00055 | 9564..10064 | + | 501 | WP_012774852 | TrbH protein | - |
| HTT42_RS00060 | 10071..11492 | + | 1422 | WP_012774853 | TrbI/VirB10 family protein | virB10 |
| HTT42_RS00065 | 11510..12292 | + | 783 | WP_012774854 | P-type conjugative transfer protein TrbJ | virB5 |
| HTT42_RS00070 | 12298..12525 | + | 228 | WP_086959606 | entry exclusion lipoprotein TrbK | - |
| HTT42_RS00075 | 12535..14217 | + | 1683 | WP_169628992 | P-type conjugative transfer protein TrbL | virB6 |
| HTT42_RS00080 | 14234..14731 | + | 498 | WP_086959604 | conjugal transfer protein TraL | - |
| HTT42_RS00085 | 14743..15357 | + | 615 | WP_012774857 | transglycosylase SLT domain-containing protein | - |
| HTT42_RS00090 | 15368..16096 | + | 729 | WP_165761827 | TraX family protein | - |
| HTT42_RS00095 | 16174..16566 | + | 393 | WP_237306416 | OmpA family protein | - |
| HTT42_RS00100 | 16586..16852 | + | 267 | WP_169628993 | hypothetical protein | - |
| HTT42_RS00105 | 16902..17411 | + | 510 | WP_172692985 | hypothetical protein | - |
| HTT42_RS00110 | 17484..17879 | + | 396 | WP_172692986 | hypothetical protein | - |
| HTT42_RS00115 | 17886..18548 | + | 663 | WP_169628996 | nuclease-related domain-containing protein | - |
| HTT42_RS00120 | 18545..19186 | - | 642 | WP_012774862 | recombinase family protein | - |
Host bacterium
| ID | 3766 | GenBank | NZ_MH548371 |
| Plasmid name | pVb1636 | Incompatibility group | - |
| Plasmid size | 56917 bp | Coordinate of oriT [Strand] | 47591..47696 [+] |
| Host baterium | Vibrio alginolyticus strain Vb1636 |
Cargo genes
| Drug resistance gene | blaCTX-M-14 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |