Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103300 |
| Name | oriT_pUCLA3 |
| Organism | Klebsiella pneumoniae |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KY940546 (49325..49372 [-], 48 nt) |
| oriT length | 48 nt |
| IRs (inverted repeats) | 5..12, 15..22 (GCAAAATT..AATTTTGC) |
| Location of nic site | 31..32 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 48 nt
>oriT_pUCLA3
ATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
ATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file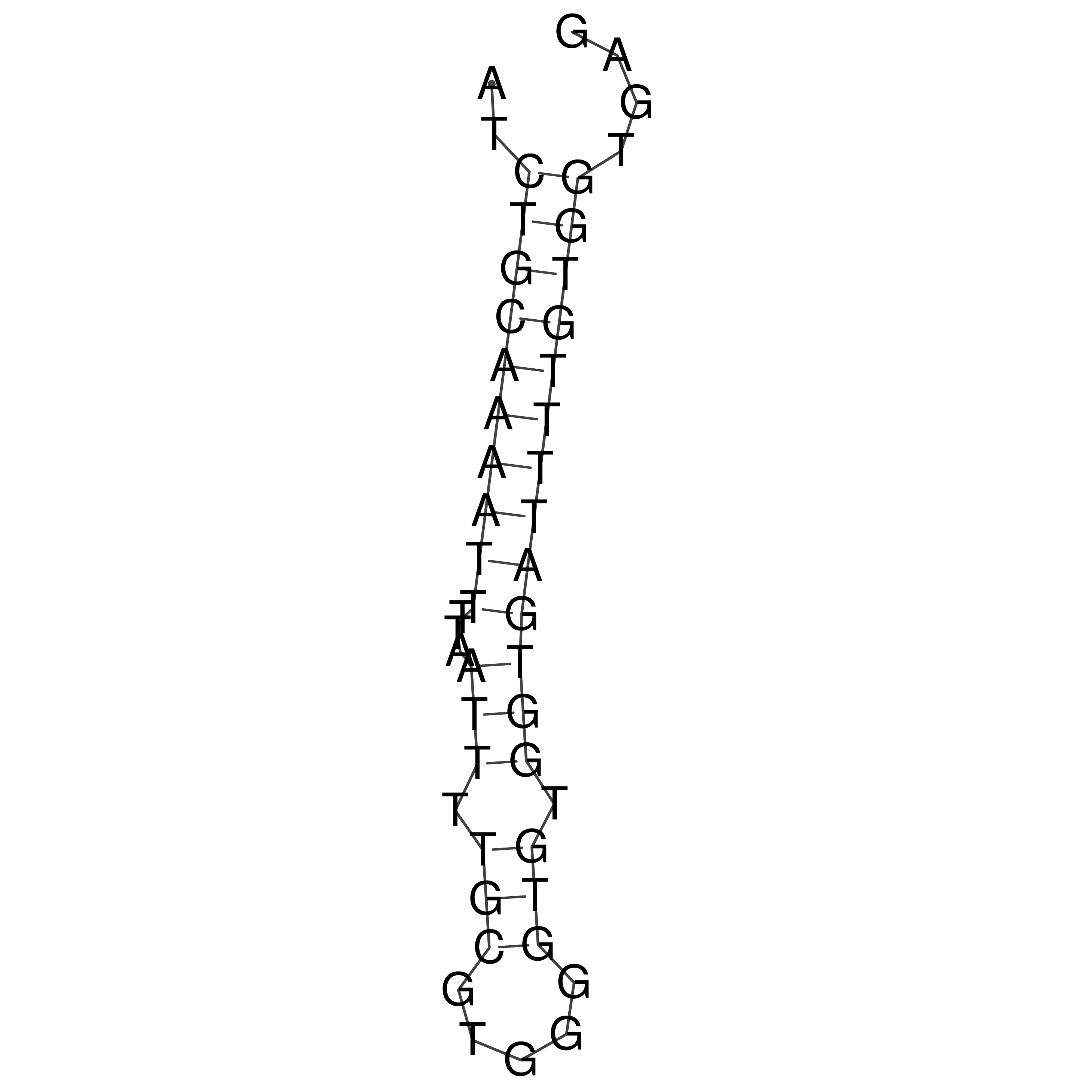
Relaxase
| ID | 2466 | GenBank | WP_020315134 |
| Name | traI_HTM92_RS00505_pUCLA3 |
UniProt ID | A0A3P2ILZ7 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190299.85 Da Isoelectric Point: 5.9174
>WP_020315134.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMEERWQGKGAEALGLDGKAVDKAVFTELLKGKLPDGSDLTRM
QDGENKHRPGYDLTFSAPKSVSMLAMLGGDKRLVDAHNQAVTEAVKQLETLTATRVMADGKSETVLTGNL
IVAKFNHDTNRNQEPQLHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYANQTAFGKLYREALKPLVE
KLGYETEVVGKHGMWEMKDVPVEAFSSRSREVREAAGPDASLKSRDVAALDTRRSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAEEIARAPVAPEKTDGPDITDVVTRAIAGLSDRKVQFTYADVLARTVGQLE
AKPGVFELARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEIQRQNHVSVTPDASVVREAP
FSDAVSVLAQDRPAMGIVAGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSREFLAGDARLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGKQATADIISEPDKSARYSRLAQEFAVSVREGQESVAQISGTREQNVLNNVIRDTLKNEGALGSRD
MMVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFVIDRVTASSNMLTLKDREGERLDLKVSAV
DSQWTLFRTNKLPVAEGERLAVLGKIPDTRLKGGESVTVLKAEAGQLTVQRPGQKTTQTLPVGSSPFDGI
KVGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHIRLYSAQDAARTTEKLARHTAFSVVSE
QLKVRSGEADLEAAIAQQKAGLHTPAEQAIHLSVPLLESNSLTFTRPQLLATALETGGGKVPMKDIEATI
QAQIKAGSLLNVPVASGHGNDLLISRQTWDAEKSVLTRVLEGKDAVAPLMGRVPGSLMTDLTAGQRAATR
MILETPDRFTVVQGYAGVGKTTQFKAVMSAISLLPEETRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLH
DTQLQQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIDRDISRALSTIEQVTPEQVPRKDGAWVPENSVTEFTPAQEKAIQ
DALKEGKEIPPGTPVSLHDAIVKDYTGRTPAAQASTLIITHLNADRRALNGLIHDARVQNGEVSKEEVTL
SVLVTSNIRDGELRKLSTWSEHQGAVALLDNTYQRIESVDKENQLITLKDNEGNIRLLSPREASAEGVTL
YRPEHITVSQGDRMRFSKSDPERGYVANSVWEVKTVSGDSVTLSDGKTTRTLNPKADEAQRHIDLAYAIT
AHGAQGASEPFAIALEGVEGRRKALVSFESAYVALSRMKQHAQVYTDNREGWTTAVSRSQEKASAHDVLE
PQNTRAVKNAGQLYGRAQPLDETAAGRAALQQSGLARGRSPGKFISPGKKYPQPHVALPAFDKNGKEAGI
WLSPLTDRDGRLEAIGGEGRIMGDETARFVALQNSRNGESLLAGNMGEGVRTARDNPDSGVIVRLAGDDR
PWNPEAITGGRIWADPLPNTPLPDTGTDIPLPPEVLAQRAAEEAQRREMEKQAEQAAREVAGEEKKAGEP
GDRIKEVIGDVIRGLERDRPGEEKTPLTDDPQTRRQEAAIQQVASESLQRERLQEMERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMEERWQGKGAEALGLDGKAVDKAVFTELLKGKLPDGSDLTRM
QDGENKHRPGYDLTFSAPKSVSMLAMLGGDKRLVDAHNQAVTEAVKQLETLTATRVMADGKSETVLTGNL
IVAKFNHDTNRNQEPQLHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYANQTAFGKLYREALKPLVE
KLGYETEVVGKHGMWEMKDVPVEAFSSRSREVREAAGPDASLKSRDVAALDTRRSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAEEIARAPVAPEKTDGPDITDVVTRAIAGLSDRKVQFTYADVLARTVGQLE
AKPGVFELARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEIQRQNHVSVTPDASVVREAP
FSDAVSVLAQDRPAMGIVAGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSREFLAGDARLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGKQATADIISEPDKSARYSRLAQEFAVSVREGQESVAQISGTREQNVLNNVIRDTLKNEGALGSRD
MMVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFVIDRVTASSNMLTLKDREGERLDLKVSAV
DSQWTLFRTNKLPVAEGERLAVLGKIPDTRLKGGESVTVLKAEAGQLTVQRPGQKTTQTLPVGSSPFDGI
KVGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHIRLYSAQDAARTTEKLARHTAFSVVSE
QLKVRSGEADLEAAIAQQKAGLHTPAEQAIHLSVPLLESNSLTFTRPQLLATALETGGGKVPMKDIEATI
QAQIKAGSLLNVPVASGHGNDLLISRQTWDAEKSVLTRVLEGKDAVAPLMGRVPGSLMTDLTAGQRAATR
MILETPDRFTVVQGYAGVGKTTQFKAVMSAISLLPEETRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLH
DTQLQQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIDRDISRALSTIEQVTPEQVPRKDGAWVPENSVTEFTPAQEKAIQ
DALKEGKEIPPGTPVSLHDAIVKDYTGRTPAAQASTLIITHLNADRRALNGLIHDARVQNGEVSKEEVTL
SVLVTSNIRDGELRKLSTWSEHQGAVALLDNTYQRIESVDKENQLITLKDNEGNIRLLSPREASAEGVTL
YRPEHITVSQGDRMRFSKSDPERGYVANSVWEVKTVSGDSVTLSDGKTTRTLNPKADEAQRHIDLAYAIT
AHGAQGASEPFAIALEGVEGRRKALVSFESAYVALSRMKQHAQVYTDNREGWTTAVSRSQEKASAHDVLE
PQNTRAVKNAGQLYGRAQPLDETAAGRAALQQSGLARGRSPGKFISPGKKYPQPHVALPAFDKNGKEAGI
WLSPLTDRDGRLEAIGGEGRIMGDETARFVALQNSRNGESLLAGNMGEGVRTARDNPDSGVIVRLAGDDR
PWNPEAITGGRIWADPLPNTPLPDTGTDIPLPPEVLAQRAAEEAQRREMEKQAEQAAREVAGEEKKAGEP
GDRIKEVIGDVIRGLERDRPGEEKTPLTDDPQTRRQEAAIQQVASESLQRERLQEMERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2240 | GenBank | WP_020315126 |
| Name | traD_HTM92_RS00500_pUCLA3 |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 85732.61 Da Isoelectric Point: 4.9642
>WP_020315126.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRTLDTRVDTRLSALLEAREAEGSLARALFTPDAPEPEQRDADSKTSPGQPVPESAP
VSPVPVKPVSTAETLSAQSSVKAQGAPQAKVATVPLTRPAPAATPAAASATASSAAATAATTGGTEQELA
QQPAEQGQEMMPAGMNEDGEIEDMQAYDAWLADEQTQREMQRREEVNINHSHRRDEQDDIEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRTLDTRVDTRLSALLEAREAEGSLARALFTPDAPEPEQRDADSKTSPGQPVPESAP
VSPVPVKPVSTAETLSAQSSVKAQGAPQAKVATVPLTRPAPAATPAAASATASSAAATAATTGGTEQELA
QQPAEQGQEMMPAGMNEDGEIEDMQAYDAWLADEQTQREMQRREEVNINHSHRRDEQDDIEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 51823..77379
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTM92_RS00310 (PUCLA3_00161) | 47110..47340 | + | 231 | WP_020314756 | hypothetical protein | - |
| HTM92_RS00590 | 47425..47544 | + | 120 | Protein_65 | SAM-dependent DNA methyltransferase | - |
| HTM92_RS00315 | 47830..48000 | - | 171 | WP_020314758 | hypothetical protein | - |
| HTM92_RS00320 (PUCLA3_00160) | 48193..48735 | + | 543 | WP_020314754 | antirestriction protein | - |
| HTM92_RS00325 (PUCLA3_00159) | 48758..49180 | - | 423 | WP_020315406 | transglycosylase SLT domain-containing protein | - |
| HTM92_RS00330 (PUCLA3_00158) | 49673..50068 | + | 396 | WP_022644714 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HTM92_RS00335 (PUCLA3_00157) | 50240..50926 | + | 687 | WP_022644713 | transcriptional regulator TraJ family protein | - |
| HTM92_RS00340 | 51005..51385 | + | 381 | WP_032409797 | TraY domain-containing protein | - |
| HTM92_RS00345 (PUCLA3_00155) | 51444..51809 | + | 366 | WP_020314752 | type IV conjugative transfer system pilin TraA | - |
| HTM92_RS00350 (PUCLA3_00154) | 51823..52128 | + | 306 | WP_020323523 | type IV conjugative transfer system protein TraL | traL |
| HTM92_RS00355 (PUCLA3_00153) | 52148..52714 | + | 567 | WP_020323527 | type IV conjugative transfer system protein TraE | traE |
| HTM92_RS00360 (PUCLA3_00152) | 52701..53441 | + | 741 | WP_020315147 | type-F conjugative transfer system secretin TraK | traK |
| HTM92_RS00365 (PUCLA3_00151) | 53441..54862 | + | 1422 | WP_020315140 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HTM92_RS00370 (PUCLA3_00150) | 54855..55289 | + | 435 | WP_020322358 | conjugal transfer protein TraP | - |
| HTM92_RS00375 (PUCLA3_00149) | 55323..55886 | + | 564 | WP_020315143 | type IV conjugative transfer system lipoprotein TraV | traV |
| HTM92_RS00380 (PUCLA3_00148) | 56031..56219 | + | 189 | WP_000957857 | hypothetical protein | - |
| HTM92_RS00385 (PUCLA3_00147) | 56229..57428 | + | 1200 | WP_000948429 | IS91 family transposase | - |
| HTM92_RS00390 (PUCLA3_00146) | 57721..57924 | + | 204 | WP_020315141 | hypothetical protein | - |
| HTM92_RS00395 | 58165..58578 | + | 414 | WP_020315139 | hypothetical protein | - |
| HTM92_RS00400 (PUCLA3_00145) | 58607..58906 | + | 300 | WP_020315144 | hypothetical protein | - |
| HTM92_RS00405 (PUCLA3_00144) | 58923..59147 | + | 225 | WP_022644711 | hypothetical protein | - |
| HTM92_RS00410 (PUCLA3_00143) | 59144..59548 | + | 405 | WP_020323520 | hypothetical protein | - |
| HTM92_RS00415 (PUCLA3_00142) | 59545..59835 | + | 291 | WP_020323516 | hypothetical protein | - |
| HTM92_RS00420 (PUCLA3_00141) | 59843..60241 | + | 399 | WP_072156904 | hypothetical protein | - |
| HTM92_RS00425 (PUCLA3_00140) | 60313..62952 | + | 2640 | WP_020323518 | type IV secretion system protein TraC | virb4 |
| HTM92_RS00430 (PUCLA3_00139) | 62952..63341 | + | 390 | WP_020314627 | type-F conjugative transfer system protein TrbI | - |
| HTM92_RS00435 (PUCLA3_00138) | 63341..63967 | + | 627 | WP_020314628 | type-F conjugative transfer system protein TraW | traW |
| HTM92_RS00440 (PUCLA3_00137) | 64011..64970 | + | 960 | WP_029497356 | conjugal transfer pilus assembly protein TraU | traU |
| HTM92_RS00445 (PUCLA3_00136) | 64983..65621 | + | 639 | WP_020321224 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HTM92_RS00450 (PUCLA3_00135) | 65680..67635 | + | 1956 | WP_020803160 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HTM92_RS00455 (PUCLA3_00134) | 67668..67949 | + | 282 | WP_022644736 | hypothetical protein | - |
| HTM92_RS00460 (PUCLA3_00133) | 67939..68166 | + | 228 | WP_012540032 | conjugal transfer protein TrbE | - |
| HTM92_RS00465 (PUCLA3_00132) | 68177..68503 | + | 327 | WP_020323521 | hypothetical protein | - |
| HTM92_RS00470 (PUCLA3_00131) | 68524..69276 | + | 753 | WP_020323528 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HTM92_RS00475 (PUCLA3_00130) | 69287..69526 | + | 240 | WP_020315123 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HTM92_RS00480 (PUCLA3_00129) | 69498..70073 | + | 576 | WP_020315129 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HTM92_RS00485 (PUCLA3_00128) | 70060..71430 | + | 1371 | WP_020315127 | conjugal transfer pilus assembly protein TraH | traH |
| HTM92_RS00490 (PUCLA3_00127) | 71430..74309 | + | 2880 | WP_020315125 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HTM92_RS00495 (PUCLA3_00126) | 74315..74842 | + | 528 | WP_020315130 | hypothetical protein | - |
| HTM92_RS00500 (PUCLA3_00125) | 75079..77379 | + | 2301 | WP_020315126 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 3743 | GenBank | NZ_KY940546 |
| Plasmid name | pUCLA3 | Incompatibility group | IncFII |
| Plasmid size | 88127 bp | Coordinate of oriT [Strand] | 49325..49372 [-] |
| Host baterium | Klebsiella pneumoniae |
Cargo genes
| Drug resistance gene | dfrA14 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |