Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103279 |
| Name | oriT_p1106-IncFIB |
| Organism | Escherichia coli strain 1106 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MG825372 (140433..140556 [-], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 101..106, 119..124 (TTTAAT..ATTAAA) 91..99, 113..121 (AATAATGTA..TACATTATT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTTTTTGAATCATTAGCTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file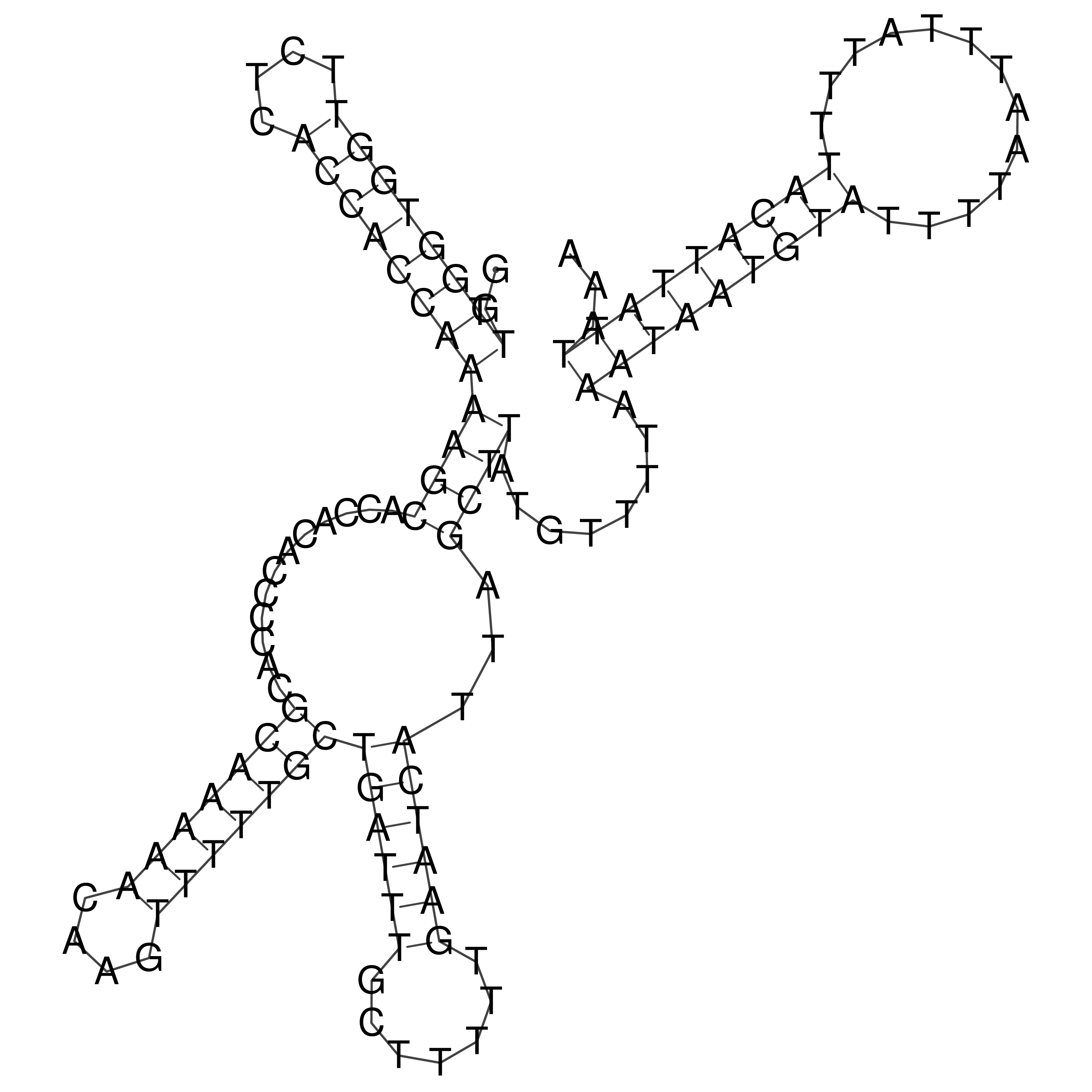
Relaxase
| ID | 2448 | GenBank | WP_172692626 |
| Name | traI_HTR02_RS00550_p1106-IncFIB |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191861.67 Da Isoelectric Point: 6.0273
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGKLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQTPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDSPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQEGMAFTPGNTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGRPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSNSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFMALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKTGLHTPAQQAIHLALPVVESKNLAFSMVDLLTEAKSFAAEGTSFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLLQRSGETPNFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWVPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDIPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIGLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKSDREVMNAERLFSTARELRDVVAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGTITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVREIAGQERDRSAISEREAALPESVLREPQRVREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1046 | GenBank | WP_001254386 |
| Name | WP_001254386_p1106-IncFIB |
UniProt ID | A0A3Z6KJB5 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9005.11 Da Isoelectric Point: 10.1422
MRRRNARGGISRTVSVYLDEDTNNRLIKAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVTEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3Z6KJB5 |
| ID | 1047 | GenBank | WP_000124981 |
| Name | WP_000124981_p1106-IncFIB |
UniProt ID | A0A630FTW9 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14542.59 Da Isoelectric Point: 5.0278
MARVNLYISNEVHEKINMIVEKRRQEGARDKDISLSGTASMLLELGLRVYDAQMERKESAFNQTEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGNPKFEYASMVDDIREKVSVEMDRFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A630FTW9 |
T4CP
| ID | 2222 | GenBank | WP_000009379 |
| Name | traD_HTR02_RS00565_p1106-IncFIB |
UniProt ID | _ |
| Length | 717 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 717 a.a. Molecular weight: 81527.16 Da Isoelectric Point: 5.1761
MSFNAKNMTQGGQIASMRIRMFSQIANIILYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIRSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLASVVALVICLITFFVVSWILGRQGKQ
QSENEITGGRQLTDNPKEVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQTRPKVAPEFIPRDINPEMENRLSAVLAAREAEGREMASLFEPDVPEVVSGEDVTQAEQPQQPVSPAI
NDKKSDAGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAVYEAWQREQNPDIQQKMQRREEVN
INVHRERGENVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2223 | GenBank | WP_072652432 |
| Name | traC_HTR02_RS00650_p1106-IncFIB |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99282.99 Da Isoelectric Point: 6.0801
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMNAAA
TQFPLPEGMNLPLTLRHYRVFFSYCSPSKKKSRADILEMENLVKIIRASLQGASITTQAVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKKNKARIDDVVDFLKNARDNDQYVESPTIRSRLDEMIVLLDQYT
ANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 114987..141128
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTR02_RS00555 (NHKPALLO_00117) | 114353..114580 | + | 228 | WP_000450520 | toxin-antitoxin system antitoxin VapB | - |
| HTR02_RS00560 (NHKPALLO_00118) | 114580..114978 | + | 399 | WP_000911333 | tRNA(fMet)-specific endonuclease VapC | - |
| HTR02_RS00565 (NHKPALLO_00119) | 114987..117140 | - | 2154 | WP_000009379 | type IV conjugative transfer system coupling protein TraD | virb4 |
| HTR02_RS00570 (NHKPALLO_00120) | 117393..118124 | - | 732 | WP_000850416 | conjugal transfer complement resistance protein TraT | - |
| HTR02_RS00575 (NHKPALLO_00121) | 118138..118647 | - | 510 | WP_000628105 | conjugal transfer entry exclusion protein TraS | - |
| HTR02_RS00580 (NHKPALLO_00122) | 118644..121469 | - | 2826 | WP_072652426 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HTR02_RS00585 (NHKPALLO_00123) | 121466..122839 | - | 1374 | WP_000944319 | conjugal transfer pilus assembly protein TraH | traH |
| HTR02_RS01030 | 122826..123218 | - | 393 | Protein_121 | F-type conjugal transfer protein TrbF | - |
| HTR02_RS00590 (NHKPALLO_00125) | 123205..123546 | - | 342 | WP_001442101 | P-type conjugative transfer protein TrbJ | - |
| HTR02_RS00595 (NHKPALLO_00126) | 123476..124021 | - | 546 | WP_000059833 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HTR02_RS00600 (NHKPALLO_00127) | 124008..124292 | - | 285 | WP_000624108 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HTR02_RS00605 (NHKPALLO_00128) | 124373..124648 | + | 276 | WP_001513501 | hypothetical protein | - |
| HTR02_RS00610 (NHKPALLO_00129) | 124650..124997 | - | 348 | WP_001287907 | conjugal transfer protein TrbA | - |
| HTR02_RS00615 (NHKPALLO_00130) | 125013..125756 | - | 744 | WP_001030378 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HTR02_RS00620 (NHKPALLO_00131) | 125749..126006 | - | 258 | WP_000864318 | conjugal transfer protein TrbE | - |
| HTR02_RS00625 (NHKPALLO_00132) | 126033..127841 | - | 1809 | WP_072645770 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HTR02_RS00630 (NHKPALLO_00133) | 127838..128476 | - | 639 | WP_000777703 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HTR02_RS00635 (NHKPALLO_00134) | 128485..129477 | - | 993 | WP_000830183 | conjugal transfer pilus assembly protein TraU | traU |
| HTR02_RS00640 (NHKPALLO_00135) | 129474..130106 | - | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| HTR02_RS00645 (NHKPALLO_00136) | 130103..130489 | - | 387 | WP_000099688 | type-F conjugative transfer system protein TrbI | - |
| HTR02_RS00650 (NHKPALLO_00137) | 130486..133113 | - | 2628 | WP_072652432 | type IV secretion system protein TraC | virb4 |
| HTR02_RS00655 (NHKPALLO_00139) | 133273..133494 | - | 222 | WP_001278690 | conjugal transfer protein TraR | - |
| HTR02_RS00660 (NHKPALLO_00140) | 133629..134144 | - | 516 | WP_000809834 | type IV conjugative transfer system lipoprotein TraV | traV |
| HTR02_RS00665 (NHKPALLO_00141) | 134141..134392 | - | 252 | WP_001038351 | conjugal transfer protein TrbG | - |
| HTR02_RS00670 (NHKPALLO_00142) | 134404..134601 | - | 198 | WP_001324648 | conjugal transfer protein TrbD | - |
| HTR02_RS00675 (NHKPALLO_00143) | 134588..135178 | - | 591 | WP_000002787 | conjugal transfer pilus-stabilizing protein TraP | - |
| HTR02_RS00680 (NHKPALLO_00144) | 135168..136595 | - | 1428 | WP_000146685 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HTR02_RS00685 (NHKPALLO_00145) | 136595..137323 | - | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| HTR02_RS00690 (NHKPALLO_00146) | 137310..137876 | - | 567 | WP_000399774 | type IV conjugative transfer system protein TraE | traE |
| HTR02_RS00695 (NHKPALLO_00147) | 137898..138209 | - | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| HTR02_RS00700 (NHKPALLO_00148) | 138224..138589 | - | 366 | WP_000340276 | type IV conjugative transfer system pilin TraA | - |
| HTR02_RS00705 (NHKPALLO_00149) | 138623..138850 | - | 228 | WP_001254386 | conjugal transfer relaxosome protein TraY | - |
| HTR02_RS00710 (NHKPALLO_00150) | 138944..139630 | - | 687 | WP_001825184 | PAS domain-containing protein | - |
| HTR02_RS00715 (NHKPALLO_00151) | 139821..140204 | - | 384 | WP_000124981 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HTR02_RS00720 (NHKPALLO_00152) | 140481..141128 | + | 648 | WP_001825185 | transglycosylase SLT domain-containing protein | virB1 |
| HTR02_RS00725 (NHKPALLO_00153) | 141425..142246 | - | 822 | WP_001234469 | DUF932 domain-containing protein | - |
| HTR02_RS01035 (NHKPALLO_00154) | 142364..142651 | - | 288 | WP_000107544 | hypothetical protein | - |
| HTR02_RS01040 | 142653..142974 | + | 322 | Protein_151 | hypothetical protein | - |
| HTR02_RS00735 (NHKPALLO_00155) | 143275..143397 | - | 123 | WP_223195199 | Hok/Gef family protein | - |
| HTR02_RS01045 (NHKPALLO_00155) | 143342..143494 | - | 153 | Protein_153 | DUF5431 family protein | - |
| HTR02_RS00740 (NHKPALLO_00156) | 143711..144430 | - | 720 | WP_001276275 | plasmid SOS inhibition protein A | - |
| HTR02_RS00745 (NHKPALLO_00157) | 144427..144861 | - | 435 | WP_000845935 | conjugation system SOS inhibitor PsiB | - |
Host bacterium
| ID | 3722 | GenBank | NZ_MG825372 |
| Plasmid name | p1106-IncFIB | Incompatibility group | IncFIB |
| Plasmid size | 190401 bp | Coordinate of oriT [Strand] | 140433..140556 [-] |
| Host baterium | Escherichia coli strain 1106 |
Cargo genes
| Drug resistance gene | sitABCD |
| Virulence gene | iroB, iroC, iroD, iroE, iroN, vat, iutA, iucC, iucB, iucA |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |