Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103265 |
| Name | oriT_p977565 |
| Organism | Escherichia coli strain 347-43491A |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KY853650 (46585..46637 [-], 53 nt) |
| oriT length | 53 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 53 nt
>oriT_p977565
CACACGATTGTAACATGACCGGAACGGTCTTGTGTACAATCGGTATCGTGCCT
CACACGATTGTAACATGACCGGAACGGTCTTGTGTACAATCGGTATCGTGCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file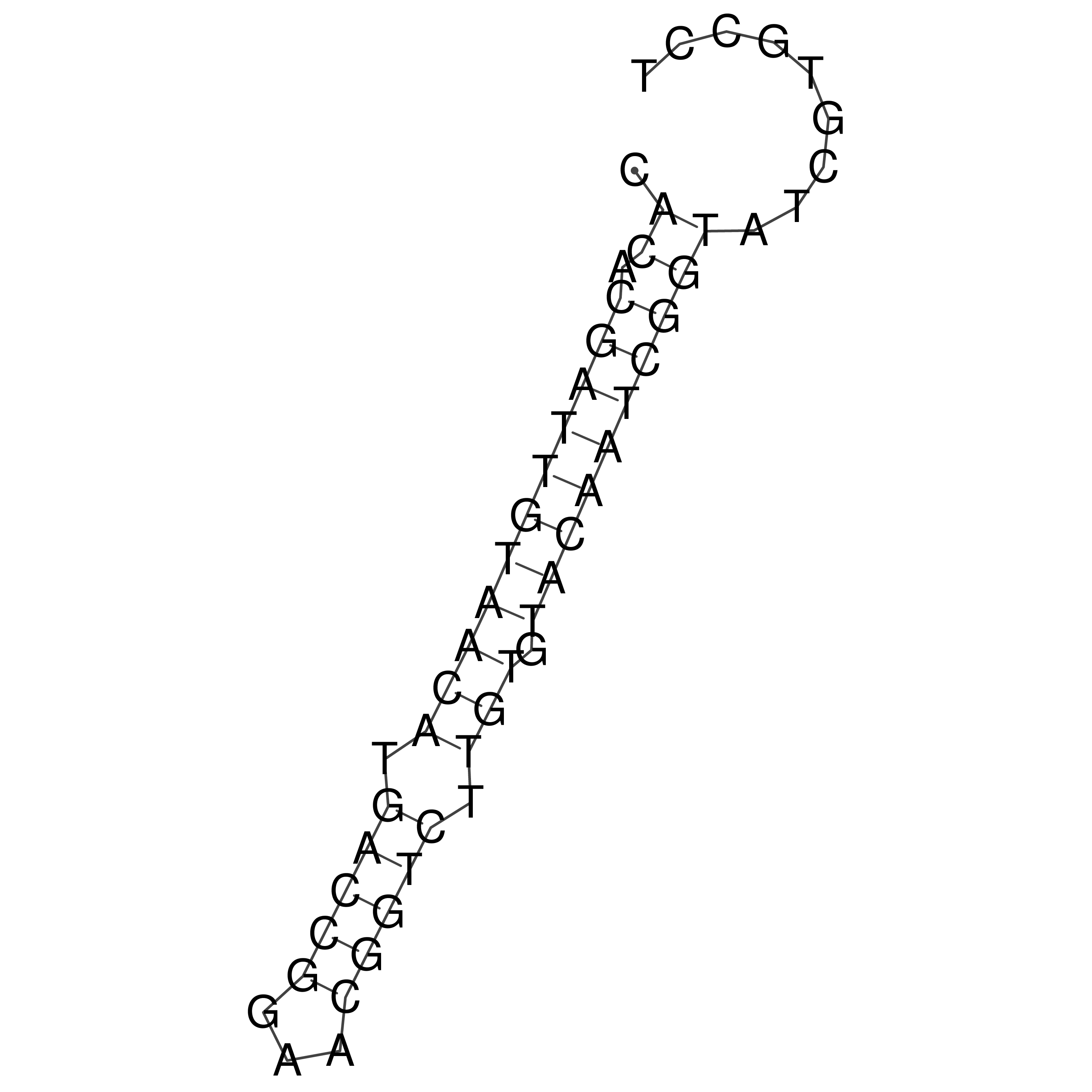
T4CP
| ID | 2209 | GenBank | WP_063268641 |
| Name | t4cp2_HTN00_RS00055_p977565 |
UniProt ID | _ |
| Length | 652 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 652 a.a. Molecular weight: 73418.87 Da Isoelectric Point: 9.2394
>WP_063268641.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacteriaceae]
MDAKKTGGLILFLLLLLVGVLIASNYLGGYTALRYSSVDMSLLKWDTFHSVISTFSGNPQYKKLVFMAWF
GFSVPLIFFAIFMLIVVIGIMPKKVIYGDARLATDMDLSKSGFFPDKKSPYKHPPILIGKMFKGRYKKQF
IYFAGQQFLILYAPTRSGKGVGIVIPNCVNYPGSMVILDIKLENWFLSAGFRQKELGQKCFLFAPAGYAE
TIDQAIKGQIRSHRWNPLDCVSRSDLLRETDLAKIAAILIPASDDPIWSDSARNLFVGLGLYLLDKERFH
LDQKAKGHNAPDVLVSISAILKTSIPDNGKDLAAWMGQEVENRSWISDKTKSFFFEFMSAPDRTRGSIKT
NFSSPLNIFSNPVTAEATNFSDFDIRDIRKKPMSIYLGLTPDALITHEKIVNLFFSLLVNENCRELPEHN
PDLKYQCLILLDEFTSMGKSEVIERAVGYTAGYNLRFVFILQNEGQGQKSDMYGQEGWTTFTENSGVVLY
YPPKSKNSLAKKISEEIGVRDMKISKRSVSSGGGNTGTSRTRNDEIIERPVLLPEEIVSLRDKKNKARNI
AIREIITSEFSRPFIANKIIWFEEPEFKRRVDIARNNHVDIPNLFTQEVMDEIAKIAEIYLPKAGGKKVM
VAGGNVITNPDLDNHDKTDVSE
MDAKKTGGLILFLLLLLVGVLIASNYLGGYTALRYSSVDMSLLKWDTFHSVISTFSGNPQYKKLVFMAWF
GFSVPLIFFAIFMLIVVIGIMPKKVIYGDARLATDMDLSKSGFFPDKKSPYKHPPILIGKMFKGRYKKQF
IYFAGQQFLILYAPTRSGKGVGIVIPNCVNYPGSMVILDIKLENWFLSAGFRQKELGQKCFLFAPAGYAE
TIDQAIKGQIRSHRWNPLDCVSRSDLLRETDLAKIAAILIPASDDPIWSDSARNLFVGLGLYLLDKERFH
LDQKAKGHNAPDVLVSISAILKTSIPDNGKDLAAWMGQEVENRSWISDKTKSFFFEFMSAPDRTRGSIKT
NFSSPLNIFSNPVTAEATNFSDFDIRDIRKKPMSIYLGLTPDALITHEKIVNLFFSLLVNENCRELPEHN
PDLKYQCLILLDEFTSMGKSEVIERAVGYTAGYNLRFVFILQNEGQGQKSDMYGQEGWTTFTENSGVVLY
YPPKSKNSLAKKISEEIGVRDMKISKRSVSSGGGNTGTSRTRNDEIIERPVLLPEEIVSLRDKKNKARNI
AIREIITSEFSRPFIANKIIWFEEPEFKRRVDIARNNHVDIPNLFTQEVMDEIAKIAEIYLPKAGGKKVM
VAGGNVITNPDLDNHDKTDVSE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2354..26403
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTN00_RS00395 | 3..329 | + | 327 | Protein_0 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HTN00_RS00005 | 326..1702 | - | 1377 | WP_000750519 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HTN00_RS00010 | 1715..2350 | - | 636 | WP_000934978 | A24 family peptidase | - |
| HTN00_RS00015 | 2354..2782 | - | 429 | WP_001326801 | lytic transglycosylase domain-containing protein | virB1 |
| HTN00_RS00020 | 2903..3460 | - | 558 | WP_097728049 | type 4 pilus major pilin | - |
| HTN00_RS00025 | 3505..4614 | - | 1110 | WP_000974900 | type II secretion system F family protein | - |
| HTN00_RS00030 | 4605..6143 | - | 1539 | WP_045146419 | ATPase, T2SS/T4P/T4SS family | virB11 |
| HTN00_RS00035 | 6168..6662 | - | 495 | WP_045146418 | type IV pilus biogenesis protein PilP | - |
| HTN00_RS00040 | 6646..7968 | - | 1323 | WP_000454142 | type 4b pilus protein PilO2 | - |
| HTN00_RS00045 | 8007..9650 | - | 1644 | WP_045146416 | PilN family type IVB pilus formation outer membrane protein | - |
| HTN00_RS00050 | 9685..10087 | - | 403 | Protein_10 | sigma 54-interacting transcriptional regulator | - |
| HTN00_RS00055 | 10134..12092 | - | 1959 | WP_063268641 | type IV secretory system conjugative DNA transfer family protein | - |
| HTN00_RS00060 | 12108..13163 | - | 1056 | WP_001059977 | P-type DNA transfer ATPase VirB11 | virB11 |
| HTN00_RS00065 | 13182..14321 | - | 1140 | WP_015057165 | TrbI/VirB10 family protein | virB10 |
| HTN00_RS00070 | 14311..15012 | - | 702 | WP_049824867 | TrbG/VirB9 family P-type conjugative transfer protein | - |
| HTN00_RS00075 | 15078..15812 | - | 735 | WP_015057166 | type IV secretion system protein | virB8 |
| HTN00_RS00080 | 15974..18331 | - | 2358 | WP_063268639 | VirB4 family type IV secretion system protein | virb4 |
| HTN00_RS00085 | 18337..18657 | - | 321 | WP_000362081 | VirB3 family type IV secretion system protein | virB3 |
| HTN00_RS00090 | 18728..19018 | - | 291 | WP_063268638 | conjugal transfer protein | - |
| HTN00_RS00095 | 19018..19602 | - | 585 | WP_063268637 | lytic transglycosylase domain-containing protein | virB1 |
| HTN00_RS00100 | 19623..20027 | - | 405 | WP_236918862 | hypothetical protein | - |
| HTN00_RS00105 | 20140..20577 | - | 438 | WP_063268636 | type IV pilus biogenesis protein PilM | - |
| HTN00_RS00110 | 20583..21818 | - | 1236 | WP_063278137 | TcpQ domain-containing protein | - |
| HTN00_RS00115 | 21821..22120 | - | 300 | WP_000835764 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| HTN00_RS00120 | 22188..22469 | - | 282 | WP_000638823 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| HTN00_RS00125 | 22459..22710 | - | 252 | WP_000121741 | hypothetical protein | - |
| HTN00_RS00130 | 22810..23445 | - | 636 | WP_015059536 | hypothetical protein | - |
| HTN00_RS00135 | 23493..24302 | + | 810 | WP_172690835 | DUF5710 domain-containing protein | - |
| HTN00_RS00140 | 24525..24749 | - | 225 | WP_000713561 | EexN family lipoprotein | - |
| HTN00_RS00145 | 24758..25402 | - | 645 | WP_001310442 | type IV secretion system protein | - |
| HTN00_RS00150 | 25408..26403 | - | 996 | WP_001028541 | type IV secretion system protein | virB6 |
| HTN00_RS00155 | 26407..26664 | - | 258 | WP_000739144 | hypothetical protein | - |
| HTN00_RS00160 | 26661..27014 | - | 354 | WP_236918887 | hypothetical protein | - |
| HTN00_RS00165 | 27226..27915 | - | 690 | WP_123873527 | hypothetical protein | - |
| HTN00_RS00170 | 27926..28096 | - | 171 | WP_000550721 | hypothetical protein | - |
| HTN00_RS00175 | 28100..28543 | - | 444 | WP_000964333 | NfeD family protein | - |
| HTN00_RS00180 | 28546..28800 | - | 255 | WP_024181945 | hypothetical protein | - |
| HTN00_RS00185 | 28923..29876 | - | 954 | WP_072815250 | SPFH domain-containing protein | - |
| HTN00_RS00190 | 29903..30079 | - | 177 | WP_045146385 | hypothetical protein | - |
| HTN00_RS00195 | 30072..30287 | - | 216 | WP_001127357 | DUF1187 family protein | - |
| HTN00_RS00200 | 30280..30786 | - | 507 | WP_001326595 | CaiF/GrlA family transcriptional regulator | - |
Host bacterium
| ID | 3708 | GenBank | NZ_KY853650 |
| Plasmid name | p977565 | Incompatibility group | IncI2 |
| Plasmid size | 63381 bp | Coordinate of oriT [Strand] | 46585..46637 [-] |
| Host baterium | Escherichia coli strain 347-43491A |
Cargo genes
| Drug resistance gene | mcr-1.11 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |