Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103212 |
| Name | oriT_FWSEC0088|unnamed2 |
| Organism | Escherichia coli strain FWSEC0088 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_RRFH01000138 (76039..76124 [+], 86 nt) |
| oriT length | 86 nt |
| IRs (inverted repeats) | 61..68, 73..80 (TTGGTGGT..ACCACCAA) 27..34, 37..44 (GCAAAAAC..GTTTTTGC) 8..13, 21..26 (TGATTT..AAATCA) |
| Location of nic site | 53..54 |
| Conserved sequence flanking the nic site |
GGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 86 nt
AATTACATGATTTAAAACGCAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAGAACCACCAACCTGTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file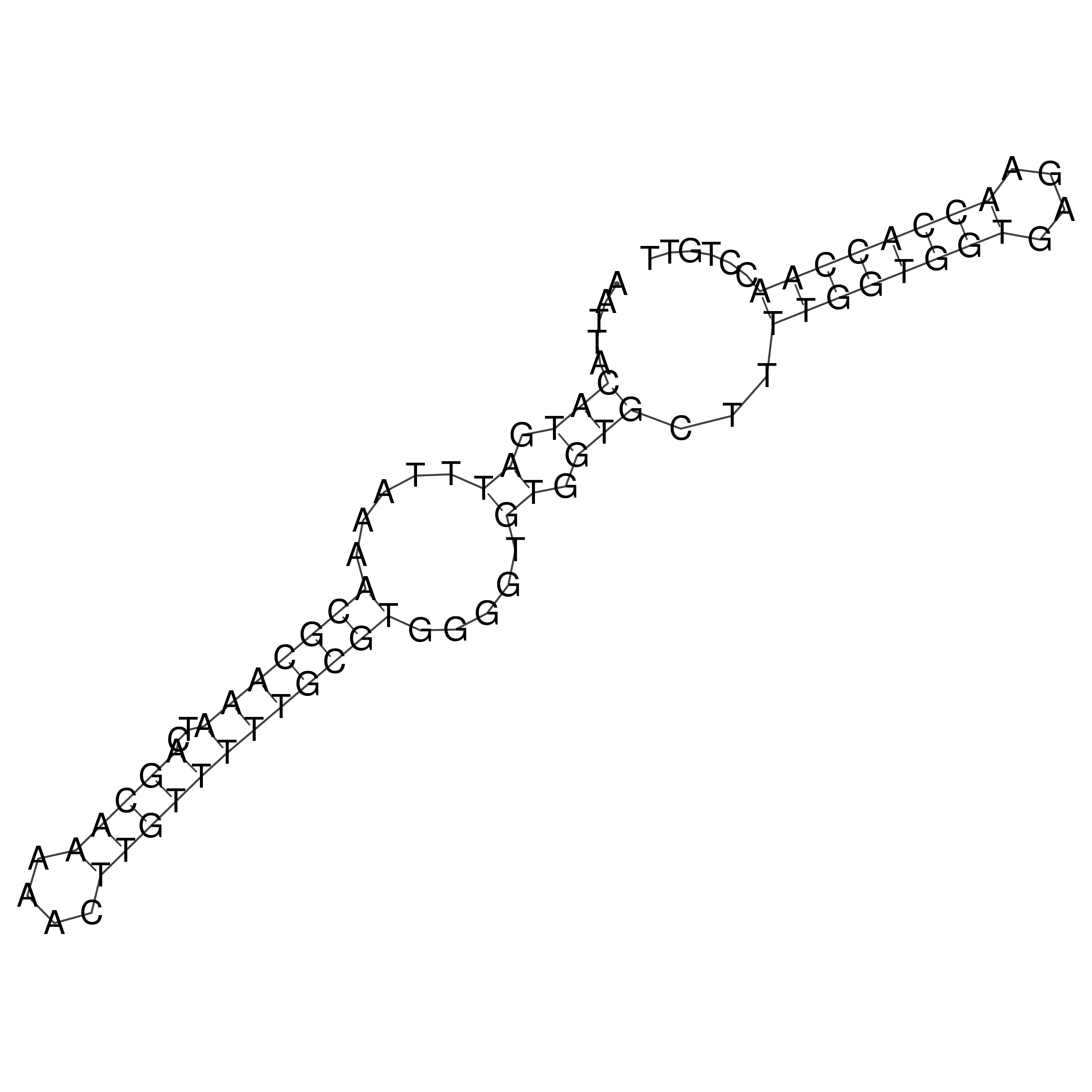
Relaxase
| ID | 2405 | GenBank | WP_033815172 |
| Name | traI_C9Z38_RS26825_FWSEC0088|unnamed2 |
UniProt ID | A0A0P7RUT6 |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191751.37 Da Isoelectric Point: 5.7364
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMLT
LKETGFDIRAYRDAADQRAETRTQTPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVSRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKISLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKIPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPATLPVSDSPFTALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTSFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMETLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVKMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVVIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDVREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLEDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVMAVSGDSVTLSDGQQTRVIRPGQEQAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAISKAVQKGTAHDVL
EPGADREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVWIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAEEKPDQP
DGKTEQAVRAIAGQEQDRASSSERETALPESVLRESQRVQDAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1018 | GenBank | WP_000089265 |
| Name | WP_000089265_FWSEC0088|unnamed2 |
UniProt ID | A0A1E5M428 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 8472.68 Da Isoelectric Point: 9.9218
MSRNIIRPAPGNKVLLVLDDATNHKLLGARERSGRTKTNEVLVRLRDHLSRFPDFYNLDAIKEGTGETDS
IIKDL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1E5M428 |
| ID | 1019 | GenBank | WP_033802152 |
| Name | WP_033802152_FWSEC0088|unnamed2 |
UniProt ID | A0A0P7P9U0 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14535.47 Da Isoelectric Point: 4.7116
MARVILYISNDVYDKVNAIVEQRRQEGARDKDISVSGTASMLLELGLRVYEAQMERKESAFNQTEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSSELERFFQKNDEE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A0P7P9U0 |
T4CP
| ID | 2166 | GenBank | WP_044809095 |
| Name | traD_C9Z38_RS26830_FWSEC0088|unnamed2 |
UniProt ID | _ |
| Length | 735 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 735 a.a. Molecular weight: 83686.34 Da Isoelectric Point: 5.1711
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIRSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLASVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVSTGKSEVIRRLANYAR
KRGDMVVIYDRSCEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNVANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTYLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHSGESFTIRDWMRGVREDKQNGWLFISSNADTHASLKPVVSMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVLNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKEMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPEVASGEDVTQAEQPQQPQQPQQPQQ
PQQPQQPQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMTAYEAWQ
QENHPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2167 | GenBank | WP_136769903 |
| Name | traC_C9Z38_RS26950_FWSEC0088|unnamed2 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 99550.31 Da Isoelectric Point: 6.0813
MSNNPLEAVTQAVNSLVTALKLPDESAKANEILGEMSFPQFSRLLPYRDYNQESGLFMNDTTLGFMLEAI
PINGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAA
ETQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGAYITTQTVDAQAFIDIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRKSTARILNFHLARNPEIAF
LWNMADNYSNLLNPEMSISCPFILTLALVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLTRQNRARIDDVVDFLKNARDNDQYAESPTIRSRLDEMIVLLDQY
TANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGW
RLLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKY
NQLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEG
LSIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 46341..76692
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| C9Z38_RS26830 (C9Z38_26845) | 46341..48548 | - | 2208 | WP_044809095 | type IV conjugative transfer system coupling protein TraD | virb4 |
| C9Z38_RS26835 (C9Z38_26850) | 48604..49335 | - | 732 | WP_054626809 | hypothetical protein | - |
| C9Z38_RS26840 (C9Z38_26855) | 49503..49763 | - | 261 | WP_001077900 | hypothetical protein | - |
| C9Z38_RS26845 (C9Z38_26860) | 49823..50554 | - | 732 | WP_033814940 | conjugal transfer complement resistance protein TraT | - |
| C9Z38_RS26850 (C9Z38_26865) | 50586..51083 | - | 498 | WP_033802149 | entry exclusion protein | - |
| C9Z38_RS26855 (C9Z38_26870) | 51099..53921 | - | 2823 | WP_033802129 | conjugal transfer mating-pair stabilization protein TraG | traG |
| C9Z38_RS26860 (C9Z38_26875) | 53918..55291 | - | 1374 | WP_033802130 | conjugal transfer pilus assembly protein TraH | traH |
| C9Z38_RS26865 (C9Z38_26880) | 55278..55670 | - | 393 | WP_000662210 | F-type conjugal transfer protein TrbF | - |
| C9Z38_RS26870 (C9Z38_26885) | 55624..55938 | - | 315 | WP_001244902 | P-type conjugative transfer protein TrbJ | - |
| C9Z38_RS26875 (C9Z38_26890) | 55928..56482 | - | 555 | WP_000059845 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| C9Z38_RS26880 (C9Z38_26895) | 56469..56750 | - | 282 | WP_000049684 | type-F conjugative transfer system pilin chaperone TraQ | - |
| C9Z38_RS26885 (C9Z38_26900) | 56816..57217 | - | 402 | WP_225394595 | hypothetical protein | - |
| C9Z38_RS26890 (C9Z38_26905) | 57348..57713 | - | 366 | WP_001009441 | hypothetical protein | - |
| C9Z38_RS26895 (C9Z38_26910) | 57727..58470 | - | 744 | WP_001030408 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| C9Z38_RS26900 (C9Z38_26915) | 58463..58720 | - | 258 | WP_000864350 | conjugal transfer protein TrbE | - |
| C9Z38_RS26905 (C9Z38_26920) | 58734..60584 | - | 1851 | WP_033802133 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| C9Z38_RS26910 (C9Z38_26925) | 60581..61003 | - | 423 | WP_001229308 | HNH endonuclease signature motif containing protein | - |
| C9Z38_RS26915 (C9Z38_26930) | 61029..61400 | - | 372 | WP_000056336 | hypothetical protein | - |
| C9Z38_RS26920 (C9Z38_26935) | 61397..62035 | - | 639 | WP_000087538 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| C9Z38_RS26925 (C9Z38_26940) | 62062..62583 | - | 522 | WP_000505571 | hypothetical protein | - |
| C9Z38_RS26935 (C9Z38_26950) | 62988..63980 | - | 993 | WP_032210964 | conjugal transfer pilus assembly protein TraU | traU |
| C9Z38_RS26940 (C9Z38_26955) | 63977..64609 | - | 633 | WP_021521304 | type-F conjugative transfer system protein TraW | traW |
| C9Z38_RS26945 (C9Z38_26960) | 64606..64992 | - | 387 | WP_021521305 | type-F conjugative transfer system protein TrbI | - |
| C9Z38_RS26950 (C9Z38_26965) | 64989..67619 | - | 2631 | WP_136769903 | type IV secretion system protein TraC | virb4 |
| C9Z38_RS26955 (C9Z38_26970) | 67746..68093 | - | 348 | WP_000802783 | hypothetical protein | - |
| C9Z38_RS26965 (C9Z38_26980) | 68278..68496 | - | 219 | WP_033802135 | hypothetical protein | - |
| C9Z38_RS26970 (C9Z38_26985) | 68576..69052 | - | 477 | WP_032210967 | hypothetical protein | - |
| C9Z38_RS26975 (C9Z38_26990) | 69045..69266 | - | 222 | WP_001278689 | conjugal transfer protein TraR | - |
| C9Z38_RS26980 (C9Z38_26995) | 69401..69916 | - | 516 | WP_033802138 | type IV conjugative transfer system lipoprotein TraV | traV |
| C9Z38_RS26985 (C9Z38_27000) | 69913..70185 | - | 273 | WP_033802139 | conjugal transfer protein TrbD | - |
| C9Z38_RS26990 (C9Z38_27005) | 70124..70684 | - | 561 | WP_000232572 | conjugal transfer pilus-stabilizing protein TraP | - |
| C9Z38_RS26995 (C9Z38_27010) | 70674..72128 | - | 1455 | WP_032210972 | F-type conjugal transfer pilus assembly protein TraB | traB |
| C9Z38_RS27000 (C9Z38_27015) | 72128..72856 | - | 729 | WP_032210974 | type-F conjugative transfer system secretin TraK | traK |
| C9Z38_RS27005 (C9Z38_27020) | 72843..73409 | - | 567 | WP_033802141 | type IV conjugative transfer system protein TraE | traE |
| C9Z38_RS27010 (C9Z38_27025) | 73431..73742 | - | 312 | WP_033802142 | type IV conjugative transfer system protein TraL | traL |
| C9Z38_RS27015 (C9Z38_27030) | 73757..74122 | - | 366 | WP_000991522 | type IV conjugative transfer system pilin TraA | - |
| C9Z38_RS27020 (C9Z38_27035) | 74155..74382 | - | 228 | WP_000089265 | conjugal transfer relaxosome protein TraY | - |
| C9Z38_RS27025 (C9Z38_27040) | 74519..75190 | - | 672 | WP_054626810 | conjugal transfer transcriptional regulator TraJ | - |
| C9Z38_RS27030 (C9Z38_27045) | 75384..75767 | - | 384 | WP_033802152 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| C9Z38_RS27040 (C9Z38_27055) | 76102..76692 | + | 591 | WP_284150284 | transglycosylase SLT domain-containing protein | virB1 |
| C9Z38_RS27045 (C9Z38_27060) | 76990..77811 | - | 822 | WP_001562881 | DUF932 domain-containing protein | - |
| C9Z38_RS27050 (C9Z38_27065) | 77930..78217 | - | 288 | WP_000107543 | hypothetical protein | - |
| C9Z38_RS27060 (C9Z38_27075) | 78373..78675 | - | 303 | WP_001272235 | hypothetical protein | - |
| C9Z38_RS27955 | 78975..79148 | + | 174 | Protein_94 | hypothetical protein | - |
Host bacterium
| ID | 3655 | GenBank | NZ_RRFH01000138 |
| Plasmid name | FWSEC0088|unnamed2 | Incompatibility group | ColRNAI |
| Plasmid size | 79212 bp | Coordinate of oriT [Strand] | 76039..76124 [+] |
| Host baterium | Escherichia coli strain FWSEC0088 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | senB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |