Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103191 |
| Name | oriT_pSI_16ESS0300 |
| Organism | Salmonella sp. strain 16ESS0300 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_ON960348 (53825..54078 [+], 254 nt) |
| oriT length | 254 nt |
| IRs (inverted repeats) | 152..157, 159..164 (AAAAGT..ACTTTT) 121..129, 134..142 (TTAAGGCTT..AAGCCTTAA) 50..55, 59..64 (AATTTT..AAAATT) |
| Location of nic site | 77..78 |
| Conserved sequence flanking the nic site |
TTTGGTTAAA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 254 nt
>oriT_pSI_16ESS0300
GGATTTAGGTTTTTTTTTAATCGCTTCACATTTCGTTAGCATGCGAAGAAATTTTGATAAAATTCTGGTCAGTTTGGTTAAAAAGTGTTACAAGTAAGCGGTGTGGTTGAAGGGATAGATTTAAGGCTTATTCAAGCCTTAAGAAAATACTAAAAGTTACTTTTCACCCTACCGAACACCTAACAAAAAATCCATGTTGAAGATTTGAACAATTGTAATGGCGCAAGGACAATCAGCACATGTCAGAATCTGAT
GGATTTAGGTTTTTTTTTAATCGCTTCACATTTCGTTAGCATGCGAAGAAATTTTGATAAAATTCTGGTCAGTTTGGTTAAAAAGTGTTACAAGTAAGCGGTGTGGTTGAAGGGATAGATTTAAGGCTTATTCAAGCCTTAAGAAAATACTAAAAGTTACTTTTCACCCTACCGAACACCTAACAAAAAATCCATGTTGAAGATTTGAACAATTGTAATGGCGCAAGGACAATCAGCACATGTCAGAATCTGAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file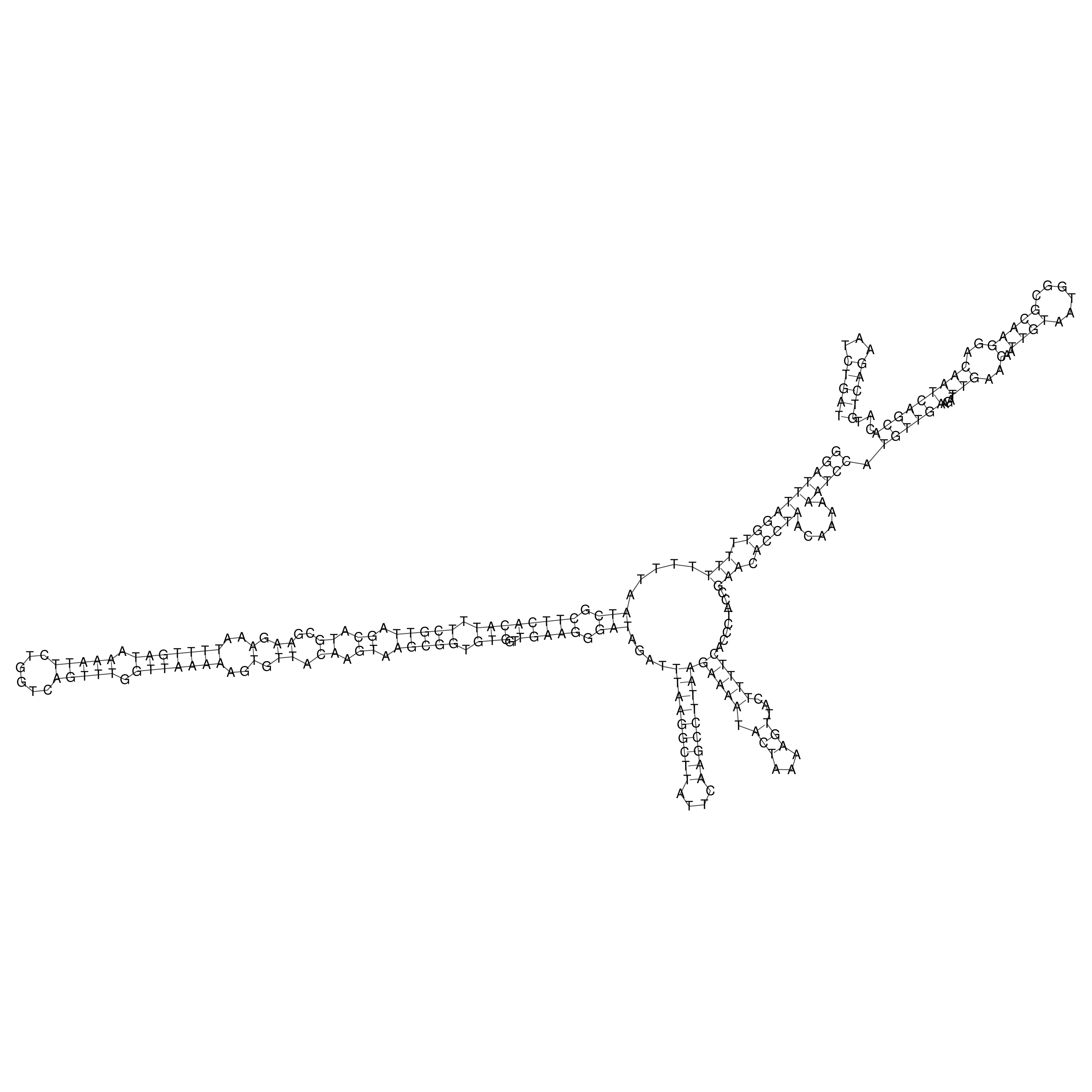
Relaxase
| ID | 2389 | GenBank | WP_001011151 |
| Name | TraI_2_PR009_RS00300_pSI_16ESS0300 |
UniProt ID | A0A1P8VU63 |
| Length | 1048 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1048 a.a. Molecular weight: 117773.04 Da Isoelectric Point: 4.6054
>WP_001011151.1 MULTISPECIES: MobH family relaxase [Enterobacterales]
MNFRALYLCIKRILGIFSSQENDATSVMIEDISSLSPFAQILGDQKYTVPDHPNPEVLKFIEYPTRPTGI
QTFNEQSILSLYREKLHSISMMLAISDSDIRDDAYTFTNLVLKPLVEYVRWIHLLPASENHHHNGIGGLL
SHSLEVAILSLKNAHHSELRPIGYQDEEVVRRKVYLYAAFICGLVHDAGKVYDLDIVSLNLASPIIWTPS
SQSLLDWARENDVVEYEIHWRKRIHNQHNIWSSVFLERILNPVCLAFLDRVNKERVYSKMITALNVYTDG
NDFLSKCVRTADFYSTGTDLNVLRDPIMGLRSNDAAARAISTIKHNFTSININNYNAKPMHIIIVNGEVY
LNENAFLDFVLNDFELHKYNFPQGEAGKTVLVESLVQRGYVEPYDDERVVHYFIPGIYSENEISNIFRNG
IGKLEFYNLLKLRWIGLIFDSYKIPDSVPGLFSVNANKDFIYIDEQKTVTEYRRPVPGRDAITKITDTVE
TAVLKVNDLGRSTASIDVDIHSKKNEGSSDDFEKKAESDNEIDNDTQIVKSEGEEAADPVIPDIEESEDE
SAKDTESHVLVNQLHELLLSAPLSNDYIVCVDAVPYLNIDATMALLPGLDEKAFSEEPYFQLTFREGSLD
GMWIVRDIDDLRLVQLGDNCAGFQLTYHEPRRPTTLKSLFNTSMYQALVINDESSVENSAPRPKQTLELP
PPRVNAVEEHSGDVEYHGTDSASATGPLKTEAVEYEHYQHLFEEEDEEHEIIDYTDFTQLSVSRPEVGSC
ATSSSVHNEKLLPEPSELPELNREQNADPQGTIERSMDVSVGQENSEPDTEGNCPPPAEVVYSQTEAAAT
SVMASEEPALPPVLEESNGEHAPTDAKGHHLSPALARLFAPTAPVEKQNPKRNRNKSSDKAEVQKPASPV
SGHNLNSKVFATTESDQNGEFSLISEGDVTELEFVEIASVLHQILSKMEVAFKRKRKNRFMVSTPNTLYL
TQSCVEKFGSQLEAQDLFNKLPQYLVNSGAVINTKCHAFNMPTLLAASDRAKVDIERIINNLKEAGNL
MNFRALYLCIKRILGIFSSQENDATSVMIEDISSLSPFAQILGDQKYTVPDHPNPEVLKFIEYPTRPTGI
QTFNEQSILSLYREKLHSISMMLAISDSDIRDDAYTFTNLVLKPLVEYVRWIHLLPASENHHHNGIGGLL
SHSLEVAILSLKNAHHSELRPIGYQDEEVVRRKVYLYAAFICGLVHDAGKVYDLDIVSLNLASPIIWTPS
SQSLLDWARENDVVEYEIHWRKRIHNQHNIWSSVFLERILNPVCLAFLDRVNKERVYSKMITALNVYTDG
NDFLSKCVRTADFYSTGTDLNVLRDPIMGLRSNDAAARAISTIKHNFTSININNYNAKPMHIIIVNGEVY
LNENAFLDFVLNDFELHKYNFPQGEAGKTVLVESLVQRGYVEPYDDERVVHYFIPGIYSENEISNIFRNG
IGKLEFYNLLKLRWIGLIFDSYKIPDSVPGLFSVNANKDFIYIDEQKTVTEYRRPVPGRDAITKITDTVE
TAVLKVNDLGRSTASIDVDIHSKKNEGSSDDFEKKAESDNEIDNDTQIVKSEGEEAADPVIPDIEESEDE
SAKDTESHVLVNQLHELLLSAPLSNDYIVCVDAVPYLNIDATMALLPGLDEKAFSEEPYFQLTFREGSLD
GMWIVRDIDDLRLVQLGDNCAGFQLTYHEPRRPTTLKSLFNTSMYQALVINDESSVENSAPRPKQTLELP
PPRVNAVEEHSGDVEYHGTDSASATGPLKTEAVEYEHYQHLFEEEDEEHEIIDYTDFTQLSVSRPEVGSC
ATSSSVHNEKLLPEPSELPELNREQNADPQGTIERSMDVSVGQENSEPDTEGNCPPPAEVVYSQTEAAAT
SVMASEEPALPPVLEESNGEHAPTDAKGHHLSPALARLFAPTAPVEKQNPKRNRNKSSDKAEVQKPASPV
SGHNLNSKVFATTESDQNGEFSLISEGDVTELEFVEIASVLHQILSKMEVAFKRKRKNRFMVSTPNTLYL
TQSCVEKFGSQLEAQDLFNKLPQYLVNSGAVINTKCHAFNMPTLLAASDRAKVDIERIINNLKEAGNL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1P8VU63 |
T4CP
| ID | 2143 | GenBank | WP_046788497 |
| Name | traD_PR009_RS00295_pSI_16ESS0300 |
UniProt ID | _ |
| Length | 694 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 694 a.a. Molecular weight: 78171.95 Da Isoelectric Point: 8.0177
>WP_046788497.1 MULTISPECIES: conjugative transfer system coupling protein TraD [Enterobacterales]
MSDSKRTNLHAQENFYRPILEYRSASILLICSVSMLYMGLSSDGLDIAPIVLFTSILLFLLCLYRCKTAA
PFLMAHWRVFKRHFMFVSLDSLRVINKSNFFSNERKYRQLVQDYQNKNKDIPERKSYFCDGFEWGPEHAD
RAYQIANLSSDKREIELPFVFNPIKRHFDAMARKMGGSNAIFAVERREPIFVTEDNWFGHTLITGNVGTG
KTVLQRLLSISMLHLGHVVVVIDPKNDAEWRESLMEEAKTLGLPFYKFHPGQPASSVCIDVCNTYTNVSD
LTSRLLSLVTVPGEVNPFVQYAKALVSNVISGLSYIEKKPSIYLIHKNMKSHMSIVNLTVKVMESCYARY
YGYDVWTEKVKYVANETLPVRFKRLAEWFTAHFMNYEGSEQIDWLDTVSQLIDYSMSDPEHMAKMTAGIM
PVFDMLIEKPLNELLSPNPNSVSSREIVTSEGMFSTGGVLYISLDGLSNPDTAAAISQLIMSDLTSCAGS
RYNAQDGDMSANSRISIFVDEAHSAINNPMINLLAQGRAAKIALFICTQTISDFIAAASVETANRITGLC
NNYISLRVNDTPTQTLVVENFGKSAISTNMVTYTTGSETSLPHNNFSGSISERKQTTLEESIPKDLLGQV
PMFHIVARLQDGRKVVGQIPIAVAEKQMKPNTTLSEMLFKKAGKVTLRQNLDIKNLNKFLRKFH
MSDSKRTNLHAQENFYRPILEYRSASILLICSVSMLYMGLSSDGLDIAPIVLFTSILLFLLCLYRCKTAA
PFLMAHWRVFKRHFMFVSLDSLRVINKSNFFSNERKYRQLVQDYQNKNKDIPERKSYFCDGFEWGPEHAD
RAYQIANLSSDKREIELPFVFNPIKRHFDAMARKMGGSNAIFAVERREPIFVTEDNWFGHTLITGNVGTG
KTVLQRLLSISMLHLGHVVVVIDPKNDAEWRESLMEEAKTLGLPFYKFHPGQPASSVCIDVCNTYTNVSD
LTSRLLSLVTVPGEVNPFVQYAKALVSNVISGLSYIEKKPSIYLIHKNMKSHMSIVNLTVKVMESCYARY
YGYDVWTEKVKYVANETLPVRFKRLAEWFTAHFMNYEGSEQIDWLDTVSQLIDYSMSDPEHMAKMTAGIM
PVFDMLIEKPLNELLSPNPNSVSSREIVTSEGMFSTGGVLYISLDGLSNPDTAAAISQLIMSDLTSCAGS
RYNAQDGDMSANSRISIFVDEAHSAINNPMINLLAQGRAAKIALFICTQTISDFIAAASVETANRITGLC
NNYISLRVNDTPTQTLVVENFGKSAISTNMVTYTTGSETSLPHNNFSGSISERKQTTLEESIPKDLLGQV
PMFHIVARLQDGRKVVGQIPIAVAEKQMKPNTTLSEMLFKKAGKVTLRQNLDIKNLNKFLRKFH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 231818..241025
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PR009_RS01260 (CPHELBEB_04308) | 231818..234499 | - | 2682 | WP_000387412 | TraC family protein | virb4 |
| PR009_RS01265 (CPHELBEB_04309) | 234508..235458 | - | 951 | WP_001022587 | IncHI-type conjugal transfer lipoprotein TrhV | traV |
| PR009_RS01270 (CPHELBEB_04310) | 235468..236220 | - | 753 | WP_183079335 | protein-disulfide isomerase HtdT | - |
| PR009_RS01275 (CPHELBEB_04311) | 236339..236821 | - | 483 | WP_000377633 | hypothetical protein | - |
| PR009_RS01280 (CPHELBEB_04312) | 236829..238184 | - | 1356 | WP_000351841 | IncHI-type conjugal transfer protein TrhB | traB |
| PR009_RS01285 (CPHELBEB_04313) | 238174..238635 | - | 462 | WP_000521240 | plasmid transfer protein HtdO | - |
| PR009_RS01290 (CPHELBEB_04314) | 238637..239908 | - | 1272 | WP_000592090 | type-F conjugative transfer system secretin TraK | traK |
| PR009_RS01295 (CPHELBEB_04315) | 239908..240696 | - | 789 | WP_000783153 | TraE/TraK family type IV conjugative transfer system protein | traE |
| PR009_RS01300 (CPHELBEB_04316) | 240708..241025 | - | 318 | WP_000043357 | type IV conjugative transfer system protein TraL | traL |
| PR009_RS01305 (CPHELBEB_04317) | 241076..241429 | - | 354 | WP_000423602 | hypothetical protein | - |
| PR009_RS01310 (CPHELBEB_04320) | 242578..243453 | - | 876 | WP_000594612 | RepB family plasmid replication initiator protein | - |
| PR009_RS01315 (CPHELBEB_04321) | 243930..244985 | + | 1056 | WP_001065779 | hypothetical protein | - |
| PR009_RS01320 (CPHELBEB_04322) | 245003..245203 | + | 201 | WP_000594931 | hypothetical protein | - |
| PR009_RS01325 (CPHELBEB_04323) | 245200..245916 | + | 717 | WP_046788421 | zinc finger-like domain-containing protein | - |
Host bacterium
| ID | 3634 | GenBank | NZ_ON960348 |
| Plasmid name | pSI_16ESS0300 | Incompatibility group | - |
| Plasmid size | 258199 bp | Coordinate of oriT [Strand] | 53825..54078 [+] |
| Host baterium | Salmonella sp. strain 16ESS0300 |
Cargo genes
| Drug resistance gene | ant(3'')-Ia, sul3, aph(3')-Ia, sul1, oqxA, oqxB, cmlA1, aadA2, dfrA12, blaCTX-M-14, fosA3, aac(3)-IVa, sul2, floR, mph(A), mcr-1.1 |
| Virulence gene | - |
| Metal resistance gene | terW, terZ, terA, terB, terC, terD, terE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |