Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103178 |
| Name | oriT_ZJ623|unnamed |
| Organism | Escherichia coli strain ZJ623 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KX580713 (44181..44283 [+], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | 64..70, 73..79 (CCACCGT..ACGGTGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 103 nt
>oriT_ZJ623|unnamed
AGCCTCGCAGAGCAGGATTCCCGTTGAGCAGCCCAGCTGCGAATAAGGGAAGGTAAAGTTGGACCACCGTTTACGGTGGGCCTACTTTACATATCCTGCCCGG
AGCCTCGCAGAGCAGGATTCCCGTTGAGCAGCCCAGCTGCGAATAAGGGAAGGTAAAGTTGGACCACCGTTTACGGTGGGCCTACTTTACATATCCTGCCCGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file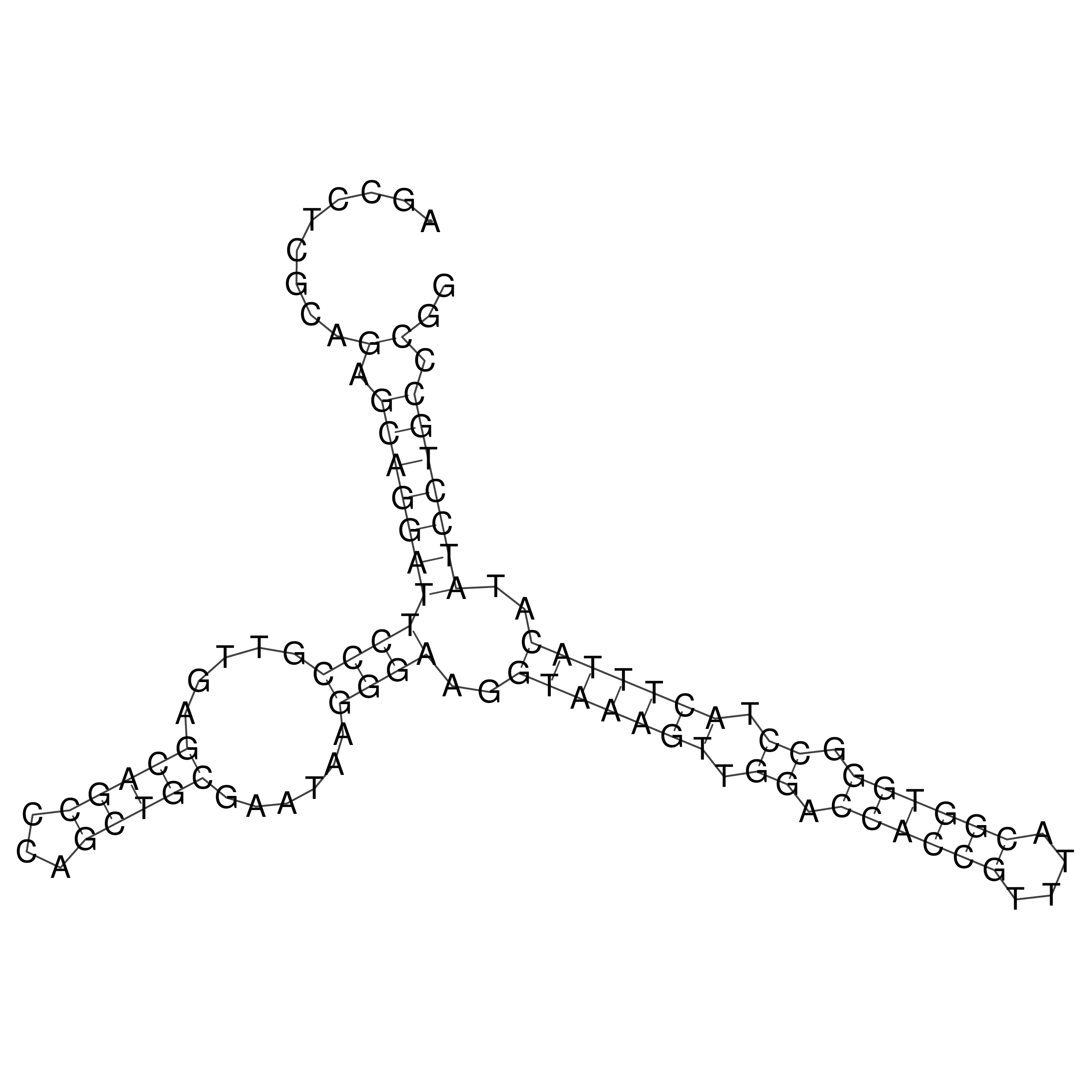
Relaxase
| ID | 2379 | GenBank | WP_050580356 |
| Name | Relaxase_HTJ09_RS00270_ZJ623|unnamed |
UniProt ID | _ |
| Length | 869 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 869 a.a. Molecular weight: 96507.42 Da Isoelectric Point: 10.4435
>WP_050580356.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Enterobacteriaceae]
MKNIKKSSFSGLADYITNPQSKQERVGVIKATNCQSQDVQDAVTEVNITQRLNTRSESDKTYHLVISFRP
GEKPSEDVLNAIEESICQSLGFGDHQRLSAVHHDTDNLHIHVAINKINPDTLTIHNPYNDYRVLAESCAY
LEKKYGLEVDNHMPTRSASENRAKDMERHSGIQSLLSWVKAECAEQLLNANSWQELHQAMRQNGLELRQK
GNGFVIAGPDGLAVKASSVDRSLSRAALEKKLGSFESSIPDTKPSRPSPEAIKQRFGSAANVKRPGDRPP
PMRENRQVHLGDVPVMQINSGKRYESKPLGSQGANTTELYGRYQSEQQRLTAARNEALRLAKVKRDRLIE
NAKRSGRLKRATIKLLKGPGVNKRYLYGLASKALGEELERANAIYKKDRQAAYDIHKRRTWADWLQFKAA
GGDDEALKTLRQRNAQQARKEANILHGDKVKKDGRIPGLKPDSVTKEGTIIYRVGESAIRDGGKRLDVSR
DAGDDGLHAALLMAKHRYGDKLTVNGSFEFKKRIVAVAAARELDVSFADPELEKLRQILTKKAAREREYI
HIPFEKRGKARQAGAMWDTEKKSWFVGPYATRDRIENFIRMNEEKDNEQRRQRYAGTERTDAGRERSGNG
NATGANEPGVTGRSTSADAGNGYGTSSSRRRTGTGDAVTTAGSQPGRSDDTRTGRSRAAEPNGERGHSKP
NVARPGRQPPPTAKNRLRAMSELGMVRIAGGSEVLLQGNVSDQLEHKTAERADQLRRDMAGAGGITTPAE
VKKPLSSAEKYINEREQKRLKGFDIPKHRGYNSTDKGLFEYAGQRKSGLETLALLKRDDEIVVMPVSDYT
ARRLSNLKVGESVTLTPSGSVRTTKGRSR
MKNIKKSSFSGLADYITNPQSKQERVGVIKATNCQSQDVQDAVTEVNITQRLNTRSESDKTYHLVISFRP
GEKPSEDVLNAIEESICQSLGFGDHQRLSAVHHDTDNLHIHVAINKINPDTLTIHNPYNDYRVLAESCAY
LEKKYGLEVDNHMPTRSASENRAKDMERHSGIQSLLSWVKAECAEQLLNANSWQELHQAMRQNGLELRQK
GNGFVIAGPDGLAVKASSVDRSLSRAALEKKLGSFESSIPDTKPSRPSPEAIKQRFGSAANVKRPGDRPP
PMRENRQVHLGDVPVMQINSGKRYESKPLGSQGANTTELYGRYQSEQQRLTAARNEALRLAKVKRDRLIE
NAKRSGRLKRATIKLLKGPGVNKRYLYGLASKALGEELERANAIYKKDRQAAYDIHKRRTWADWLQFKAA
GGDDEALKTLRQRNAQQARKEANILHGDKVKKDGRIPGLKPDSVTKEGTIIYRVGESAIRDGGKRLDVSR
DAGDDGLHAALLMAKHRYGDKLTVNGSFEFKKRIVAVAAARELDVSFADPELEKLRQILTKKAAREREYI
HIPFEKRGKARQAGAMWDTEKKSWFVGPYATRDRIENFIRMNEEKDNEQRRQRYAGTERTDAGRERSGNG
NATGANEPGVTGRSTSADAGNGYGTSSSRRRTGTGDAVTTAGSQPGRSDDTRTGRSRAAEPNGERGHSKP
NVARPGRQPPPTAKNRLRAMSELGMVRIAGGSEVLLQGNVSDQLEHKTAERADQLRRDMAGAGGITTPAE
VKKPLSSAEKYINEREQKRLKGFDIPKHRGYNSTDKGLFEYAGQRKSGLETLALLKRDDEIVVMPVSDYT
ARRLSNLKVGESVTLTPSGSVRTTKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2127 | GenBank | WP_032310210 |
| Name | t4cp2_HTJ09_RS00260_ZJ623|unnamed |
UniProt ID | _ |
| Length | 648 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 648 a.a. Molecular weight: 71711.82 Da Isoelectric Point: 8.3842
>WP_032310210.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacteriaceae]
MRKPNNAVGPQVRNKKKTARGKLIPAMAITSLAAGIQAATQYFAYSFNYQELLGPNFNHIYAPWSYFKWY
SAWHDRLPDAFFAAGSVGATVAAGGLVLTALTSMMKANSSKANEYLHGSARWADEQDIKAAGLLGNDEGV
YVGAWEDKNGQLQYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWKHSTVVADLKGELWAMTAGWRKEYA
KNLCLKFEPAAANGSVAWNPLDEIRVGTENEVGDVQNLATLIVDPDGKGLNDHWQKTSQALLVGVILHVL
YKHKNDGTPATLPYVDSIMADPERGAGELWMEMTQYGHVNGENHPVVGAAGRDMMDRPEEEAGSVLSTAK
SYLALFRDPVVARNVSESHFKIKDLMNHDDPVSLYIVTQPNDKARLRPLVRIMLNMIVRLLADKMEFERV
NNNLTLWQRFKQAFGFSVQGTKRVQTKKTYKHRLLAMIDEFPSLGKLEIIQESLAFLAGYGIKFYLICQD
INQLRSRETGYGPDETITSNCHVQNAYPPNRTETAEHLSKLTGQTTVIKEQITTSGKRASAILGGVSKTM
QEVQRPLLTVDECLRMPGPKKNAEGLITERGDMVVYVAGFPAIYGKQPLFFQDEIFSLRASVPEPKTTDR
IRAAVAANDDASNEAIAI
MRKPNNAVGPQVRNKKKTARGKLIPAMAITSLAAGIQAATQYFAYSFNYQELLGPNFNHIYAPWSYFKWY
SAWHDRLPDAFFAAGSVGATVAAGGLVLTALTSMMKANSSKANEYLHGSARWADEQDIKAAGLLGNDEGV
YVGAWEDKNGQLQYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWKHSTVVADLKGELWAMTAGWRKEYA
KNLCLKFEPAAANGSVAWNPLDEIRVGTENEVGDVQNLATLIVDPDGKGLNDHWQKTSQALLVGVILHVL
YKHKNDGTPATLPYVDSIMADPERGAGELWMEMTQYGHVNGENHPVVGAAGRDMMDRPEEEAGSVLSTAK
SYLALFRDPVVARNVSESHFKIKDLMNHDDPVSLYIVTQPNDKARLRPLVRIMLNMIVRLLADKMEFERV
NNNLTLWQRFKQAFGFSVQGTKRVQTKKTYKHRLLAMIDEFPSLGKLEIIQESLAFLAGYGIKFYLICQD
INQLRSRETGYGPDETITSNCHVQNAYPPNRTETAEHLSKLTGQTTVIKEQITTSGKRASAILGGVSKTM
QEVQRPLLTVDECLRMPGPKKNAEGLITERGDMVVYVAGFPAIYGKQPLFFQDEIFSLRASVPEPKTTDR
IRAAVAANDDASNEAIAI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 10425..20743
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTJ09_RS00050 | 5988..6185 | - | 198 | WP_048207659 | DUF905 family protein | - |
| HTJ09_RS00310 | 6275..6544 | - | 270 | WP_228719037 | hypothetical protein | - |
| HTJ09_RS00315 | 6646..6858 | - | 213 | WP_223307920 | hypothetical protein | - |
| HTJ09_RS00060 | 6855..7103 | - | 249 | WP_071443967 | antirestriction protein ArdR | - |
| HTJ09_RS00065 | 7103..7411 | - | 309 | WP_003019666 | HigA family addiction module antitoxin | - |
| HTJ09_RS00070 | 7420..7719 | - | 300 | WP_071443968 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| HTJ09_RS00075 | 8568..9446 | - | 879 | WP_048207660 | plasmid replication initiator TrfA | - |
| HTJ09_RS00325 | 9584..9763 | - | 180 | WP_306433452 | single-stranded DNA-binding protein | - |
| HTJ09_RS00085 | 9836..10240 | + | 405 | WP_048207662 | transcriptional regulator | - |
| HTJ09_RS00090 | 10425..11381 | + | 957 | WP_032932562 | P-type conjugative transfer ATPase TrbB | virB11 |
| HTJ09_RS00095 | 11495..11878 | + | 384 | WP_003019650 | TrbC/VirB2 family protein | virB2 |
| HTJ09_RS00100 | 11881..12207 | + | 327 | WP_003019648 | conjugal transfer protein TrbD | virB3 |
| HTJ09_RS00105 | 12204..14747 | + | 2544 | WP_071443970 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| HTJ09_RS00110 | 14744..15451 | + | 708 | WP_048207665 | VirB8/TrbF family protein | virB8 |
| HTJ09_RS00115 | 15467..16357 | + | 891 | WP_003019641 | P-type conjugative transfer protein TrbG | virB9 |
| HTJ09_RS00120 | 16360..16812 | + | 453 | WP_032194780 | conjugal transfer protein TrbH | - |
| HTJ09_RS00125 | 16816..18210 | + | 1395 | WP_048207667 | TrbI/VirB10 family protein | virB10 |
| HTJ09_RS00130 | 18223..18993 | + | 771 | WP_046960647 | P-type conjugative transfer protein TrbJ | virB5 |
| HTJ09_RS00135 | 19006..19179 | + | 174 | WP_024252956 | entry exclusion lipoprotein TrbK | - |
| HTJ09_RS00140 | 19184..20743 | + | 1560 | WP_048207669 | P-type conjugative transfer protein TrbL | virB6 |
| HTJ09_RS00145 | 20754..21059 | + | 306 | WP_032310194 | lipoprotein | - |
| HTJ09_RS00150 | 21075..21845 | + | 771 | WP_048207671 | conjugal transfer protein | - |
| HTJ09_RS00155 | 21862..22548 | + | 687 | WP_053065212 | TraX family protein | - |
| HTJ09_RS00160 | 23133..23924 | - | 792 | WP_172689002 | PAP2 family protein | - |
| HTJ09_RS00165 | 23972..25597 | - | 1626 | WP_049589868 | phosphoethanolamine--lipid A transferase MCR-1.1 | - |
Host bacterium
| ID | 3621 | GenBank | NZ_KX580713 |
| Plasmid name | ZJ623|unnamed | Incompatibility group | IncP1 |
| Plasmid size | 47409 bp | Coordinate of oriT [Strand] | 44181..44283 [+] |
| Host baterium | Escherichia coli strain ZJ623 |
Cargo genes
| Drug resistance gene | mcr-1.1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |