Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103170 |
| Name | oriT_pKP1814-2 |
| Organism | Klebsiella pneumoniae strain KP1814 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KX839208 (52297..52346 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pKP1814-2
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file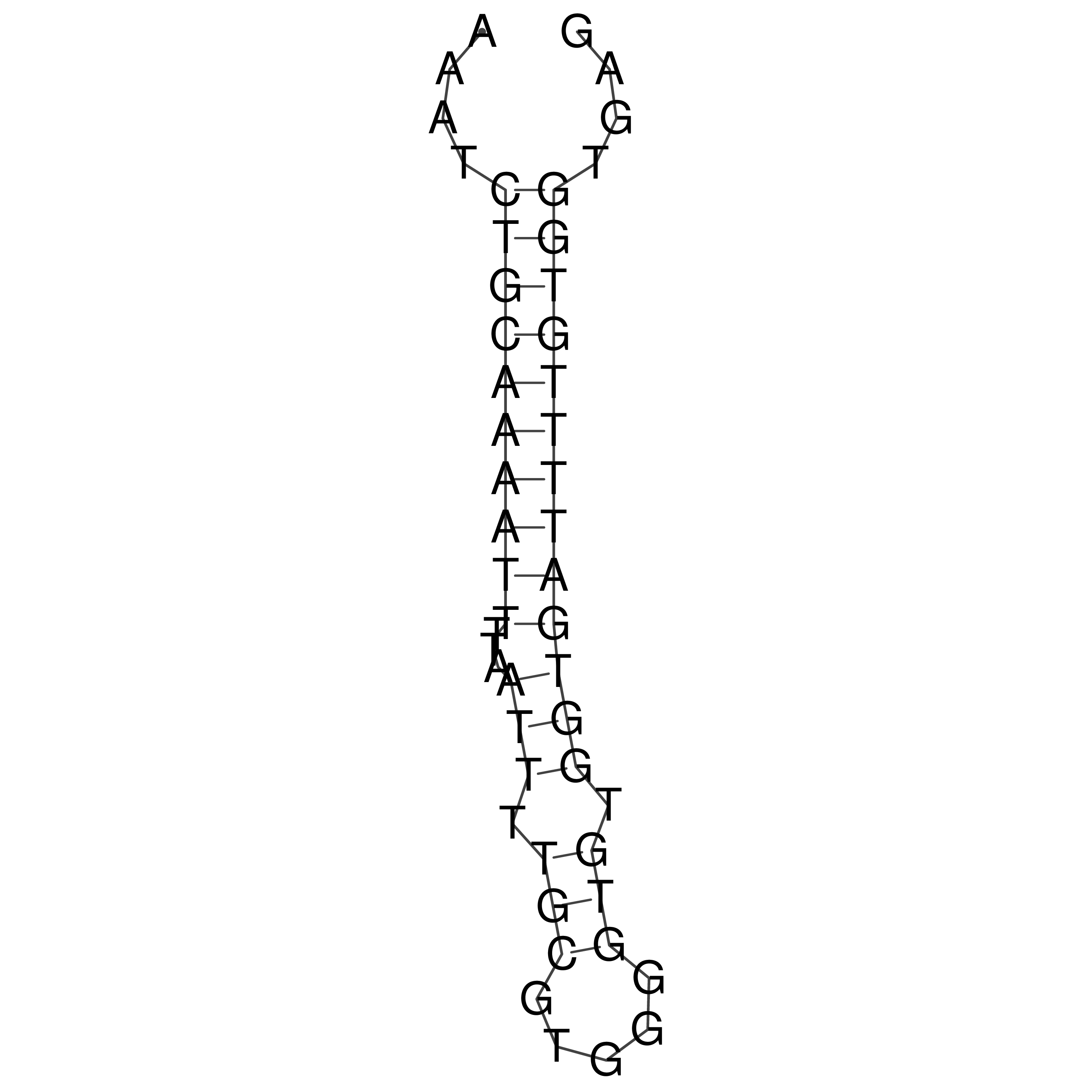
Relaxase
| ID | 2372 | GenBank | WP_023307532 |
| Name | traI_HTK02_RS00490_pKP1814-2 |
UniProt ID | A0A2R4ND60 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190476.66 Da Isoelectric Point: 6.4600
>WP_023307532.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRAMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELACAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRHNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALITIEQVTPEQVPRKEGVWAPGSSVVEFTPKQEKAIE
KALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRTLNGMIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSDGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASERYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRIWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRAMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELACAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRHNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALITIEQVTPEQVPRKEGVWAPGSSVVEFTPKQEKAIE
KALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRTLNGMIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSDGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASERYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRIWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2118 | GenBank | WP_015065643 |
| Name | traD_HTK02_RS00485_pKP1814-2 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85851.84 Da Isoelectric Point: 5.0577
>WP_015065643.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTVWCGEQLWTSIVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRRLDTRVDARLSALLEAREAEGSLARTLFMPDAPASGPADTNSHAGEQPEPISQPA
PADMTVSPAPVKAPPTTKSPAAEPSVRATEPPVLRVTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTVWCGEQLWTSIVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRRLDTRVDARLSALLEAREAEGSLARTLFMPDAPASGPADTNSHAGEQPEPISQPA
PADMTVSPAPVKAPPTTKSPAAEPSVRATEPPVLRVTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 51739..80220
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTK02_RS00260 | 46758..47108 | + | 351 | WP_023280874 | hypothetical protein | - |
| HTK02_RS00265 | 47745..48101 | + | 357 | WP_004152717 | hypothetical protein | - |
| HTK02_RS00270 | 48162..48374 | + | 213 | WP_004152718 | hypothetical protein | - |
| HTK02_RS00275 | 48385..48609 | + | 225 | WP_004152719 | hypothetical protein | - |
| HTK02_RS00280 | 48690..49010 | + | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| HTK02_RS00285 | 49000..49278 | + | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| HTK02_RS00290 | 49279..49692 | + | 414 | WP_023280875 | transcriptional regulator | - |
| HTK02_RS00295 | 50523..51350 | + | 828 | WP_023307552 | DUF932 domain-containing protein | - |
| HTK02_RS00300 | 51377..51706 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| HTK02_RS00305 | 51739..52224 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| HTK02_RS00310 | 52659..53051 | + | 393 | WP_032436755 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HTK02_RS00315 | 53267..53953 | + | 687 | WP_228717881 | transcriptional regulator TraJ family protein | - |
| HTK02_RS00320 | 54037..54408 | + | 372 | WP_004208838 | TraY domain-containing protein | - |
| HTK02_RS00325 | 54462..54830 | + | 369 | WP_023307550 | type IV conjugative transfer system pilin TraA | - |
| HTK02_RS00330 | 54844..55149 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| HTK02_RS00335 | 55169..55735 | + | 567 | WP_004152602 | type IV conjugative transfer system protein TraE | traE |
| HTK02_RS00340 | 55722..56462 | + | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| HTK02_RS00345 | 56462..57886 | + | 1425 | WP_023307549 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HTK02_RS00350 | 57879..58292 | + | 414 | Protein_75 | conjugal transfer pilus-stabilizing protein TraP | - |
| HTK02_RS00355 | 58308..58484 | + | 177 | WP_172687660 | hypothetical protein | - |
| HTK02_RS00360 | 58504..59073 | + | 570 | WP_023307547 | type IV conjugative transfer system lipoprotein TraV | traV |
| HTK02_RS00365 | 59205..59615 | + | 411 | WP_023307546 | hypothetical protein | - |
| HTK02_RS00370 | 59620..59910 | + | 291 | WP_172687661 | hypothetical protein | - |
| HTK02_RS00375 | 59934..60152 | + | 219 | WP_023307545 | hypothetical protein | - |
| HTK02_RS00380 | 60153..60470 | + | 318 | WP_023307544 | hypothetical protein | - |
| HTK02_RS00385 | 60537..60941 | + | 405 | WP_023307543 | hypothetical protein | - |
| HTK02_RS00390 | 60938..61228 | + | 291 | WP_023307542 | hypothetical protein | - |
| HTK02_RS00395 | 61236..61634 | + | 399 | WP_072158599 | hypothetical protein | - |
| HTK02_RS00400 | 61706..64345 | + | 2640 | WP_016528865 | type IV secretion system protein TraC | virb4 |
| HTK02_RS00405 | 64345..64734 | + | 390 | WP_004152591 | type-F conjugative transfer system protein TrbI | - |
| HTK02_RS00410 | 64734..65360 | + | 627 | WP_023307538 | type-F conjugative transfer system protein TraW | traW |
| HTK02_RS00415 | 65404..66363 | + | 960 | WP_015065634 | conjugal transfer pilus assembly protein TraU | traU |
| HTK02_RS00420 | 66376..67014 | + | 639 | WP_015065635 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HTK02_RS00425 | 67073..69028 | + | 1956 | WP_023307537 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HTK02_RS00430 | 69060..69296 | + | 237 | WP_023307536 | conjugal transfer protein TrbE | - |
| HTK02_RS01055 | 69293..69478 | + | 186 | WP_023307535 | hypothetical protein | - |
| HTK02_RS00435 | 69524..69850 | + | 327 | WP_004152685 | hypothetical protein | - |
| HTK02_RS00440 | 69871..70623 | + | 753 | WP_015065637 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HTK02_RS00445 | 70634..70873 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HTK02_RS00450 | 70845..71402 | + | 558 | WP_004152678 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HTK02_RS00455 | 71447..71890 | + | 444 | WP_015065638 | F-type conjugal transfer protein TrbF | - |
| HTK02_RS00460 | 71868..73247 | + | 1380 | WP_011977731 | conjugal transfer pilus assembly protein TraH | traH |
| HTK02_RS00465 | 73247..76096 | + | 2850 | WP_108479757 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HTK02_RS00470 | 76102..76677 | + | 576 | WP_064187086 | hypothetical protein | - |
| HTK02_RS00480 | 76818..77549 | + | 732 | Protein_101 | conjugal transfer complement resistance protein TraT | - |
| HTK02_RS00485 | 77911..80220 | + | 2310 | WP_015065643 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 3613 | GenBank | NZ_KX839208 |
| Plasmid name | pKP1814-2 | Incompatibility group | IncFII |
| Plasmid size | 187349 bp | Coordinate of oriT [Strand] | 52297..52346 [-] |
| Host baterium | Klebsiella pneumoniae strain KP1814 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | mrkJ, mrkF, mrkD, mrkC, mrkB, mrkA |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |