Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103133 |
| Name | oriT_pOP-I |
| Organism | Enterobacter cloacae subsp. cloacae strain MN201516 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KY126370 (50399..50485 [+], 87 nt) |
| oriT length | 87 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 87 nt
>oriT_pOP-I
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTAATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCGTCTTTCG
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTAATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCGTCTTTCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file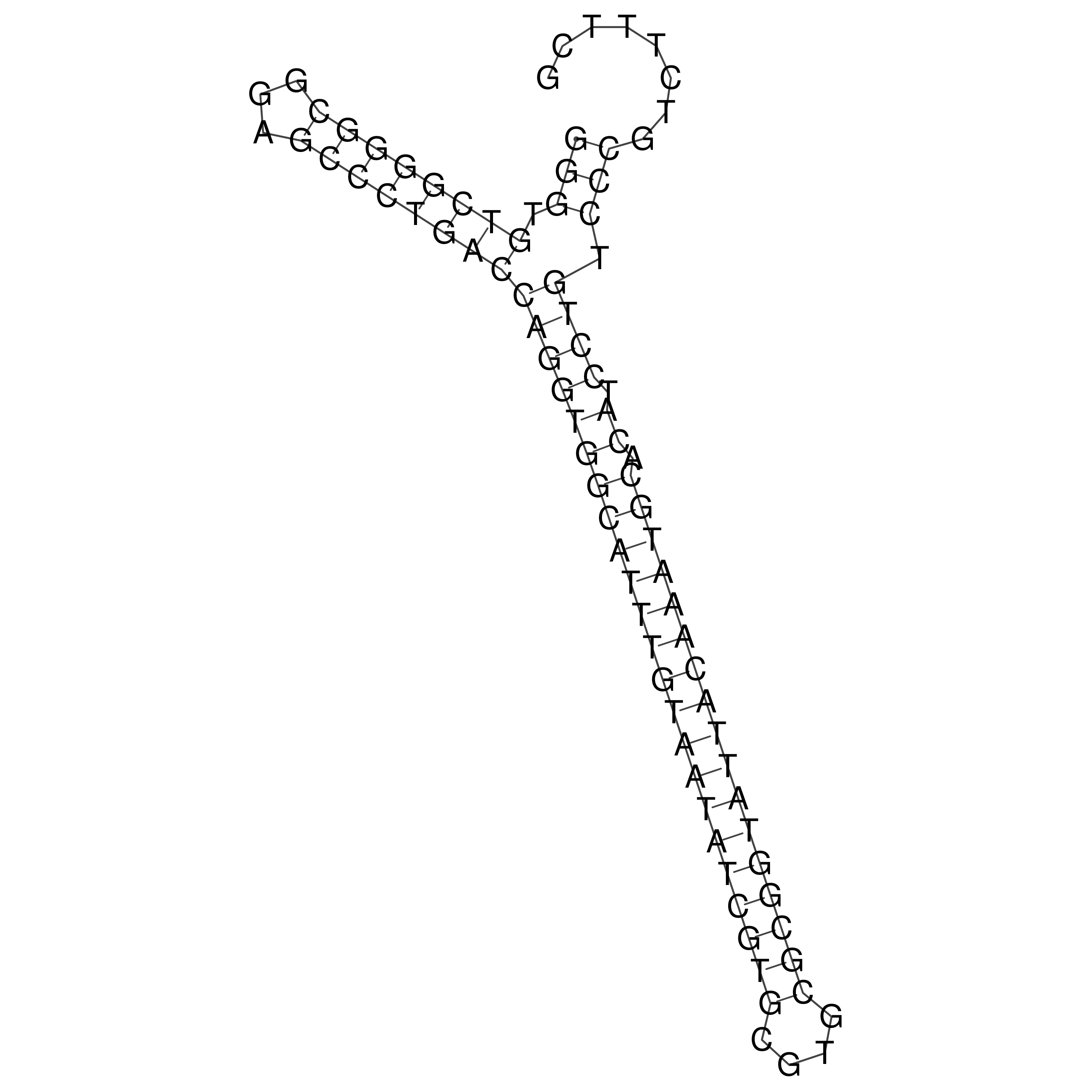
Relaxase
| ID | 2350 | GenBank | WP_172689589 |
| Name | Relaxase_HTL24_RS00220_pOP-I |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 102435.55 Da Isoelectric Point: 7.9064
>WP_172689589.1 relaxase/mobilization nuclease domain-containing protein [Enterobacter cloacae]
MIPVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMEAVKASGTKPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIESAAEEMEFSARKAKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVSHTLSRLGLVGHQFVAAVHTDTDNLHVHIAVNRVHPETGYLNRLAYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNRIVRRTSVERDRQSAWKRDRNQSLSEYIADTAIAGLRESPVTDWSSLHQRFAKEGLFLTKD
SGELKVKDGWDSERPGVTLSAFGHAFDTDKLTRKLGEFVPPAQDIFSQVPAVGRYEPDKIIMPSRPERMT
EKESLSDYATARLRASLIALDQDPTQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDPTRTPVRAERVWP
ALTKAILDSYKGGWQPVPKDIFQQVPPAEQFTGGGLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQDIEMLIGSKQFSWERCHELLARQGLLLMREQQGLVVMDAYNHEQTPVKASHVHPDLTLAR
AEPHAGSFVSVPADIFERIKPVSRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLHGRERYQVIHDECRMRKARIRIEHRDPLVRKLHYHIAELQRMQALITLKEAMKAERAELIGQGK
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYRDRRGNAVKSTTDSRCVVICEPGGTPMYSGVPGLQASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNTGDYDALKMDMVKVAPVLFSRSSTVAMAPEGNNEQFNTAFAKM
VAWHNINHRDDGEYQISRPDIDQVRVESEMRFAEMADQGRNPEHERNEPGSNWKPPSPL
MIPVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMEAVKASGTKPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIESAAEEMEFSARKAKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVSHTLSRLGLVGHQFVAAVHTDTDNLHVHIAVNRVHPETGYLNRLAYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNRIVRRTSVERDRQSAWKRDRNQSLSEYIADTAIAGLRESPVTDWSSLHQRFAKEGLFLTKD
SGELKVKDGWDSERPGVTLSAFGHAFDTDKLTRKLGEFVPPAQDIFSQVPAVGRYEPDKIIMPSRPERMT
EKESLSDYATARLRASLIALDQDPTQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDPTRTPVRAERVWP
ALTKAILDSYKGGWQPVPKDIFQQVPPAEQFTGGGLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQDIEMLIGSKQFSWERCHELLARQGLLLMREQQGLVVMDAYNHEQTPVKASHVHPDLTLAR
AEPHAGSFVSVPADIFERIKPVSRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLHGRERYQVIHDECRMRKARIRIEHRDPLVRKLHYHIAELQRMQALITLKEAMKAERAELIGQGK
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYRDRRGNAVKSTTDSRCVVICEPGGTPMYSGVPGLQASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNTGDYDALKMDMVKVAPVLFSRSSTVAMAPEGNNEQFNTAFAKM
VAWHNINHRDDGEYQISRPDIDQVRVESEMRFAEMADQGRNPEHERNEPGSNWKPPSPL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2079 | GenBank | WP_172689587 |
| Name | trbC_HTL24_RS00210_pOP-I |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 87424.66 Da Isoelectric Point: 6.5761
>WP_172689587.1 F-type conjugative transfer protein TrbC [Enterobacter cloacae]
MNNTPVNQALMKRNAWNSPVLELMRDHLLYAGIMAGSVVAGFIWPLAIPLLLLLAIVASISFGTHRWRMP
MRMPVHLNKVDPSEDRKVRRSLFRVFPSLFQYETTQESIGKGIFYMGYKRMNDIGRELWLTIDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IQNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAGLYCRAIDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHKLE
GLDSEALEITKYKGQFPYMNYLDEVGAYYTERIAVQATQVRSLEFALIMMSQDQERIENQTSAANVATLM
QNAGTKVAGKIVSDDKTARTISNAAGQEARANMVSLQRQDGIIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIESQDKTCNAPVIINRYITINPPNLKQLRKLVPRTAQRRLPAPENVSKIIG
VLTEKPSRRRKASRSEPWKIIDTFQQRLANRQEAHNLMTEYDMDHSGREENLWEEALEIIRTTTMAERTI
RYITLNKAESESTEVNAEEIKLAPDAILRTLTLPQPAYVPPARYRNEPFISPKLPGSQHPDMEWPE
MNNTPVNQALMKRNAWNSPVLELMRDHLLYAGIMAGSVVAGFIWPLAIPLLLLLAIVASISFGTHRWRMP
MRMPVHLNKVDPSEDRKVRRSLFRVFPSLFQYETTQESIGKGIFYMGYKRMNDIGRELWLTIDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IQNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAGLYCRAIDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHKLE
GLDSEALEITKYKGQFPYMNYLDEVGAYYTERIAVQATQVRSLEFALIMMSQDQERIENQTSAANVATLM
QNAGTKVAGKIVSDDKTARTISNAAGQEARANMVSLQRQDGIIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIESQDKTCNAPVIINRYITINPPNLKQLRKLVPRTAQRRLPAPENVSKIIG
VLTEKPSRRRKASRSEPWKIIDTFQQRLANRQEAHNLMTEYDMDHSGREENLWEEALEIIRTTTMAERTI
RYITLNKAESESTEVNAEEIKLAPDAILRTLTLPQPAYVPPARYRNEPFISPKLPGSQHPDMEWPE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 9830..44613
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTL24_RS00030 | 5088..5894 | + | 807 | WP_236901763 | TcpQ domain-containing protein | - |
| HTL24_RS00035 | 5891..6331 | + | 441 | WP_236901764 | type IV pilus biogenesis protein PilM | - |
| HTL24_RS00040 | 6346..8022 | + | 1677 | WP_172689563 | PilN family type IVB pilus formation outer membrane protein | - |
| HTL24_RS00045 | 8024..9328 | + | 1305 | WP_172689564 | type 4b pilus protein PilO2 | - |
| HTL24_RS00050 | 9312..9827 | + | 516 | WP_172689565 | type IV pilus biogenesis protein PilP | - |
| HTL24_RS00055 | 9830..11389 | + | 1560 | WP_172689566 | ATPase, T2SS/T4P/T4SS family | virB11 |
| HTL24_RS00060 | 11382..12476 | + | 1095 | WP_172689627 | type II secretion system F family protein | - |
| HTL24_RS00065 | 12568..13104 | + | 537 | WP_236901766 | type 4 pilus major pilin | - |
| HTL24_RS00070 | 13189..13671 | + | 483 | WP_172689628 | lytic transglycosylase domain-containing protein | virB1 |
| HTL24_RS00075 | 13656..14330 | + | 675 | WP_172689568 | prepilin peptidase | - |
| HTL24_RS00080 | 14327..15664 | + | 1338 | WP_172689629 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HTL24_RS00085 | 16527..17687 | + | 1161 | WP_048210079 | site-specific integrase | - |
| HTL24_RS00090 (MN002) | 17764..18183 | + | 420 | WP_075262562 | hypothetical protein | - |
| HTL24_RS00095 | 18329..19165 | + | 837 | WP_172689569 | hypothetical protein | traE |
| HTL24_RS00100 | 19268..20476 | + | 1209 | WP_172689570 | conjugal transfer protein TraF | - |
| HTL24_RS00105 (MN003) | 20497..21015 | + | 519 | WP_172689571 | hypothetical protein | - |
| HTL24_RS00110 | 21103..21576 | + | 474 | WP_172689572 | DotD/TraH family lipoprotein | - |
| HTL24_RS00115 | 21573..22379 | + | 807 | WP_172689573 | type IV secretory system conjugative DNA transfer family protein | traI |
| HTL24_RS00120 | 22391..23551 | + | 1161 | WP_172689574 | plasmid transfer ATPase TraJ | virB11 |
| HTL24_RS00125 | 23548..23835 | + | 288 | WP_048210072 | IcmT/TraK family protein | traK |
| HTL24_RS00130 (MN004) | 23907..27455 | + | 3549 | WP_172689575 | LPD7 domain-containing protein | - |
| HTL24_RS00135 | 27473..27823 | + | 351 | WP_048210070 | hypothetical protein | traL |
| HTL24_RS00140 | 27829..28530 | + | 702 | WP_172689576 | DotI/IcmL/TraM family protein | traM |
| HTL24_RS00145 | 28541..29524 | + | 984 | WP_172689577 | DotH/IcmK family type IV secretion protein | traN |
| HTL24_RS00150 | 29524..30840 | + | 1317 | WP_172689578 | conjugal transfer protein TraO | traO |
| HTL24_RS00155 | 30824..31594 | + | 771 | WP_172689579 | conjugal transfer protein TraP | traP |
| HTL24_RS00160 | 31594..32118 | + | 525 | WP_172689580 | conjugal transfer protein TraQ | traQ |
| HTL24_RS00165 | 32153..32557 | + | 405 | WP_172689581 | DUF6750 family protein | traR |
| HTL24_RS00170 | 32742..33473 | + | 732 | WP_236901765 | conjugal transfer protein TraT | traT |
| HTL24_RS00175 | 33554..36607 | + | 3054 | WP_172689582 | ATP-binding protein | traU |
| HTL24_RS00180 | 37189..38391 | + | 1203 | WP_048210062 | conjugal transfer protein TraW | traW |
| HTL24_RS00185 | 38388..38930 | + | 543 | WP_053070179 | conjugal transfer protein TraX | - |
| HTL24_RS00190 | 38988..41150 | + | 2163 | WP_172689583 | DotA/TraY family protein | traY |
| HTL24_RS00195 | 41211..41843 | + | 633 | WP_172689584 | plasmid IncI1-type surface exclusion protein ExcA | - |
| HTL24_RS00200 | 42156..43364 | + | 1209 | WP_172689585 | conjugal transfer protein TrbA | trbA |
| HTL24_RS00205 | 43417..44613 | + | 1197 | WP_172689586 | thioredoxin fold domain-containing protein | trbB |
| HTL24_RS00210 | 44617..46917 | + | 2301 | WP_172689587 | F-type conjugative transfer protein TrbC | - |
| HTL24_RS00215 (MN005) | 46990..47259 | + | 270 | WP_172689588 | hypothetical protein | - |
Host bacterium
| ID | 3576 | GenBank | NZ_KY126370 |
| Plasmid name | pOP-I | Incompatibility group | - |
| Plasmid size | 114969 bp | Coordinate of oriT [Strand] | 50399..50485 [+] |
| Host baterium | Enterobacter cloacae subsp. cloacae strain MN201516 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merC, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |