Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103129 |
| Name | oriT_pIMI-6 |
| Organism | Enterobacter cloacae strain N14-0444 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KX786187 (132204..132260 [-], 57 nt) |
| oriT length | 57 nt |
| IRs (inverted repeats) | 13..20, 23..30 (GCAAAATT..AATTTTGC) |
| Location of nic site | 39..40 |
| Conserved sequence flanking the nic site |
GGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 57 nt
>oriT_pIMI-6
TAAAGTTAATCAGCAAAATTTTAATTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAG
TAAAGTTAATCAGCAAAATTTTAATTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file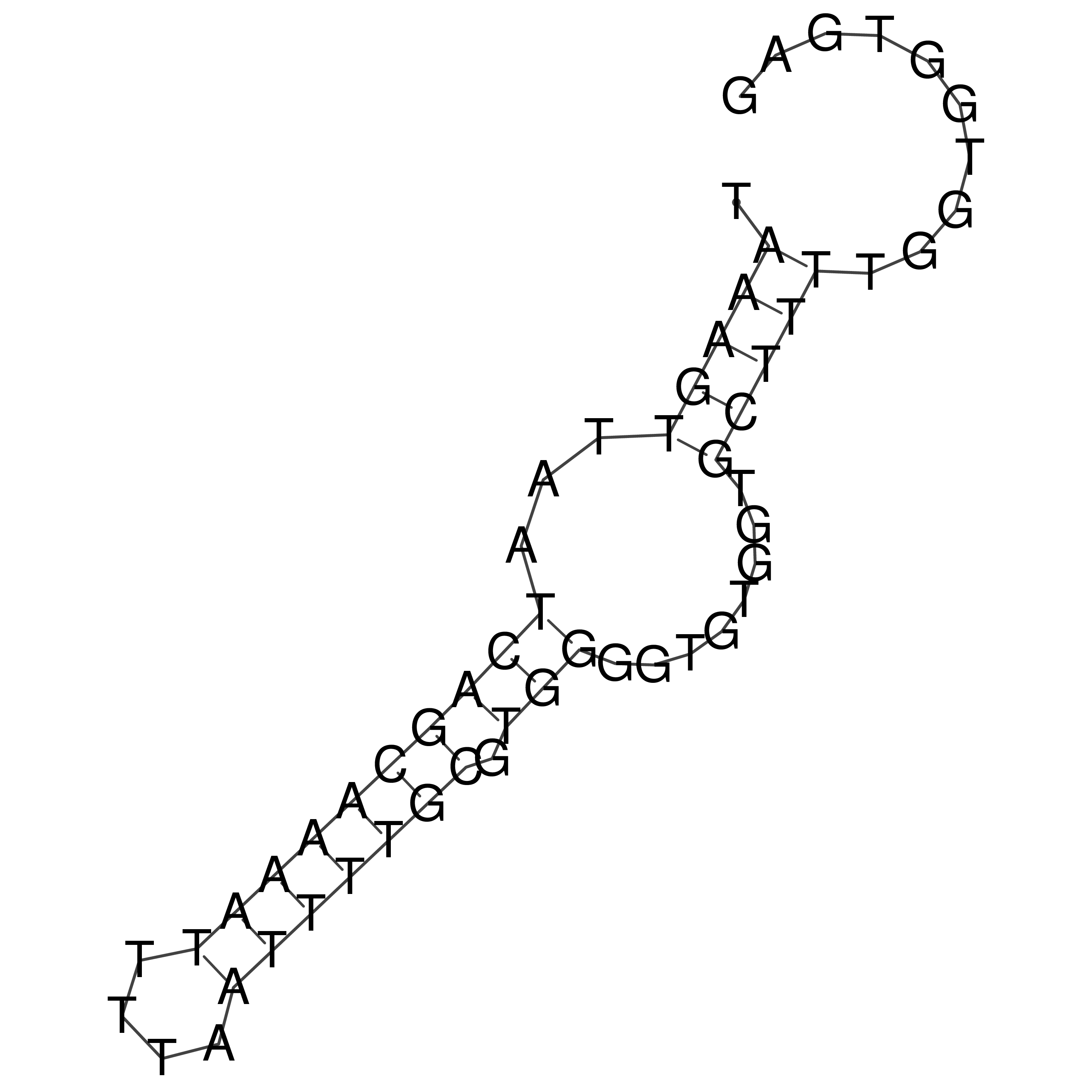
Relaxase
| ID | 2346 | GenBank | WP_172689111 |
| Name | traI_HTK44_RS00865_pIMI-6 |
UniProt ID | _ |
| Length | 1746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1746 a.a. Molecular weight: 189268.71 Da Isoelectric Point: 6.2261
>WP_172689111.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacter cloacae complex]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRIQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVDTAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTRHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVTA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMQT
LKETGFDIKAYREAADLRVVQGNIPVTAPEVIDINSSVGQSIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDLGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEARSRERFVIDRVTGRNNSLTLRNVQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLKGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVGQ
GWVESPGRSVSDSAKVFASLSQRELDNTTLNKLALSGRAVQLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQVIEKEADRQI
RSGALLSVPVSPGNGLQLLVSRQSYNAEKSILRHVLEGKEAVTPLMDKVPDAQLAGLTDGQQNATRMILG
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
HQLRGGERPDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLINRDIGSALTTIESVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDMMSGGQPATLLEAIVKDYTGRTPEAQAQTIVITALNADRRQVNAMIHEARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAVRLTATAKAQGEVPAGRAALRQAGLQPEGSMAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEVRPWN
PGAITGGRVWADGLPDGTGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRIQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVDTAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTRHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVTA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMQT
LKETGFDIKAYREAADLRVVQGNIPVTAPEVIDINSSVGQSIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDLGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEARSRERFVIDRVTGRNNSLTLRNVQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLKGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVGQ
GWVESPGRSVSDSAKVFASLSQRELDNTTLNKLALSGRAVQLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQVIEKEADRQI
RSGALLSVPVSPGNGLQLLVSRQSYNAEKSILRHVLEGKEAVTPLMDKVPDAQLAGLTDGQQNATRMILG
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
HQLRGGERPDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLINRDIGSALTTIESVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDMMSGGQPATLLEAIVKDYTGRTPEAQAQTIVITALNADRRQVNAMIHEARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAVRLTATAKAQGEVPAGRAALRQAGLQPEGSMAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEVRPWN
PGAITGGRVWADGLPDGTGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2075 | GenBank | WP_172689102 |
| Name | traC_HTK44_RS00790_pIMI-6 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 98774.39 Da Isoelectric Point: 5.5375
>WP_172689102.1 MULTISPECIES: type IV secretion system protein TraC [Enterobacter cloacae complex]
MSNPLDTVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGCNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVVQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLNDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
MSNPLDTVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGCNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVVQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLNDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2076 | GenBank | WP_172689110 |
| Name | traD_HTK44_RS00860_pIMI-6 |
UniProt ID | _ |
| Length | 731 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 731 a.a. Molecular weight: 81909.33 Da Isoelectric Point: 5.1615
>WP_172689110.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacter cloacae complex]
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGVIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQIMQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKH
QSEDEITGGRQLTDDPRAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTTDGKAEERLNAALAAREHESRSIAMLFEPDADVIAPEGGAENAGQPQQPQQPQQ
PQQPQQPAPAPRAADTTSASASVAGAAGTGGVEPELKTKAEEAEQLPPGIDESGEVVDMAAWEAWQAEQD
ELSPQERQRREEVNINVHRAPEKDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGVIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQIMQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKH
QSEDEITGGRQLTDDPRAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTTDGKAEERLNAALAAREHESRSIAMLFEPDADVIAPEGGAENAGQPQQPQQPQQ
PQQPQQPAPAPRAADTTSASASVAGAAGTGGVEPELKTKAEEAEQLPPGIDESGEVVDMAAWEAWQAEQD
ELSPQERQRREEVNINVHRAPEKDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 131656..156521
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTK44_RS00910 (PIMI6_00550) | 126993..127142 | + | 150 | Protein_141 | DUF5431 family protein | - |
| HTK44_RS00685 (PIMI6_00550) | 127084..127209 | + | 126 | WP_223540704 | type I toxin-antitoxin system Hok family toxin | - |
| HTK44_RS00690 (PIMI6_00555) | 128086..128481 | + | 396 | WP_172689094 | ammonia monooxygenase | - |
| HTK44_RS00695 (PIMI6_00560) | 128543..128836 | + | 294 | WP_172686993 | hypothetical protein | - |
| HTK44_RS00700 (PIMI6_00565) | 128883..129227 | + | 345 | WP_172686994 | hypothetical protein | - |
| HTK44_RS00705 (PIMI6_00570) | 129313..129579 | + | 267 | WP_172689095 | hypothetical protein | - |
| HTK44_RS00710 (PIMI6_00575) | 129622..130452 | + | 831 | WP_172689096 | N-6 DNA methylase | - |
| HTK44_RS00715 (PIMI6_00580) | 131079..131609 | + | 531 | WP_172689097 | antirestriction protein | - |
| HTK44_RS00720 (PIMI6_00585) | 131656..132132 | - | 477 | WP_172689098 | transglycosylase SLT domain-containing protein | virB1 |
| HTK44_RS00725 (PIMI6_00590) | 132564..132959 | + | 396 | WP_172689099 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HTK44_RS00730 (PIMI6_00595) | 133144..133848 | + | 705 | WP_071667437 | hypothetical protein | - |
| HTK44_RS00735 (PIMI6_00600) | 133957..134331 | + | 375 | WP_040218544 | TraY domain-containing protein | - |
| HTK44_RS00740 (PIMI6_00605) | 134397..134747 | + | 351 | WP_172689118 | type IV conjugative transfer system pilin TraA | - |
| HTK44_RS00745 (PIMI6_00610) | 134898..135203 | + | 306 | WP_045911931 | type IV conjugative transfer system protein TraL | traL |
| HTK44_RS00750 (PIMI6_00615) | 135218..135784 | + | 567 | WP_025759824 | type IV conjugative transfer system protein TraE | traE |
| HTK44_RS00755 (PIMI6_00620) | 135771..136514 | + | 744 | WP_172689100 | type-F conjugative transfer system secretin TraK | traK |
| HTK44_RS00760 (PIMI6_00625) | 136501..137895 | + | 1395 | WP_172689101 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HTK44_RS00765 (PIMI6_00630) | 137917..138444 | + | 528 | WP_123061130 | type IV conjugative transfer system lipoprotein TraV | traV |
| HTK44_RS00770 (PIMI6_00635) | 138564..138788 | + | 225 | WP_123061131 | TraR/DksA C4-type zinc finger protein | - |
| HTK44_RS00775 | 138792..138974 | + | 183 | WP_123061132 | hypothetical protein | virb4 |
| HTK44_RS00780 (PIMI6_00640) | 138958..139254 | + | 297 | WP_123061133 | hypothetical protein | - |
| HTK44_RS00785 (PIMI6_00645) | 139244..139810 | + | 567 | WP_045379430 | conjugal transfer pilus-stabilizing protein TraP | - |
| HTK44_RS00790 (pIMI6_00650) | 139803..142433 | + | 2631 | WP_172689102 | type IV secretion system protein TraC | virb4 |
| HTK44_RS00795 (PIMI6_00655) | 142430..142777 | + | 348 | WP_172689103 | type-F conjugative transfer system protein TrbI | - |
| HTK44_RS00800 (PIMI6_00660) | 142777..143424 | + | 648 | WP_060446831 | type-F conjugative transfer system protein TraW | traW |
| HTK44_RS00805 (PIMI6_00665) | 143427..144413 | + | 987 | WP_172689104 | conjugal transfer pilus assembly protein TraU | traU |
| HTK44_RS00810 (PIMI6_00670) | 144430..145059 | + | 630 | WP_172689105 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HTK44_RS00815 (PIMI6_00675) | 145056..146936 | + | 1881 | WP_172689106 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HTK44_RS00820 (PIMI6_00680) | 146968..147156 | + | 189 | WP_025759809 | conjugal transfer protein TrbE | - |
| HTK44_RS00825 (PIMI6_00685) | 147193..147936 | + | 744 | WP_060446849 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HTK44_RS00830 | 147953..148201 | + | 249 | WP_008324126 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HTK44_RS00835 (PIMI6_00690) | 148191..148736 | + | 546 | WP_060446834 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HTK44_RS00840 (PIMI6_00700) | 148775..150109 | + | 1335 | WP_230199841 | conjugal transfer pilus assembly protein TraH | traH |
| HTK44_RS00845 (PIMI6_00705) | 150109..152928 | + | 2820 | WP_172689107 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HTK44_RS00850 (PIMI6_00710) | 152953..153426 | + | 474 | WP_172689108 | surface exclusion protein | - |
| HTK44_RS00855 (PIMI6_00715) | 153467..154207 | + | 741 | WP_172689109 | hypothetical protein | - |
| HTK44_RS00860 (PIMI6_00720) | 154326..156521 | + | 2196 | WP_172689110 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 3572 | GenBank | NZ_KX786187 |
| Plasmid name | pIMI-6 | Incompatibility group | IncFII |
| Plasmid size | 165469 bp | Coordinate of oriT [Strand] | 132204..132260 [-] |
| Host baterium | Enterobacter cloacae strain N14-0444 |
Cargo genes
| Drug resistance gene | blaIMI-3 |
| Virulence gene | hsiB1/vipA, hsiC1/vipB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |