Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103116 |
| Name | oriT_pG12-KPC-2 |
| Organism | Klebsiella pneumoniae strain K47-25 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KU665642 (72942..72991 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pG12-KPC-2
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file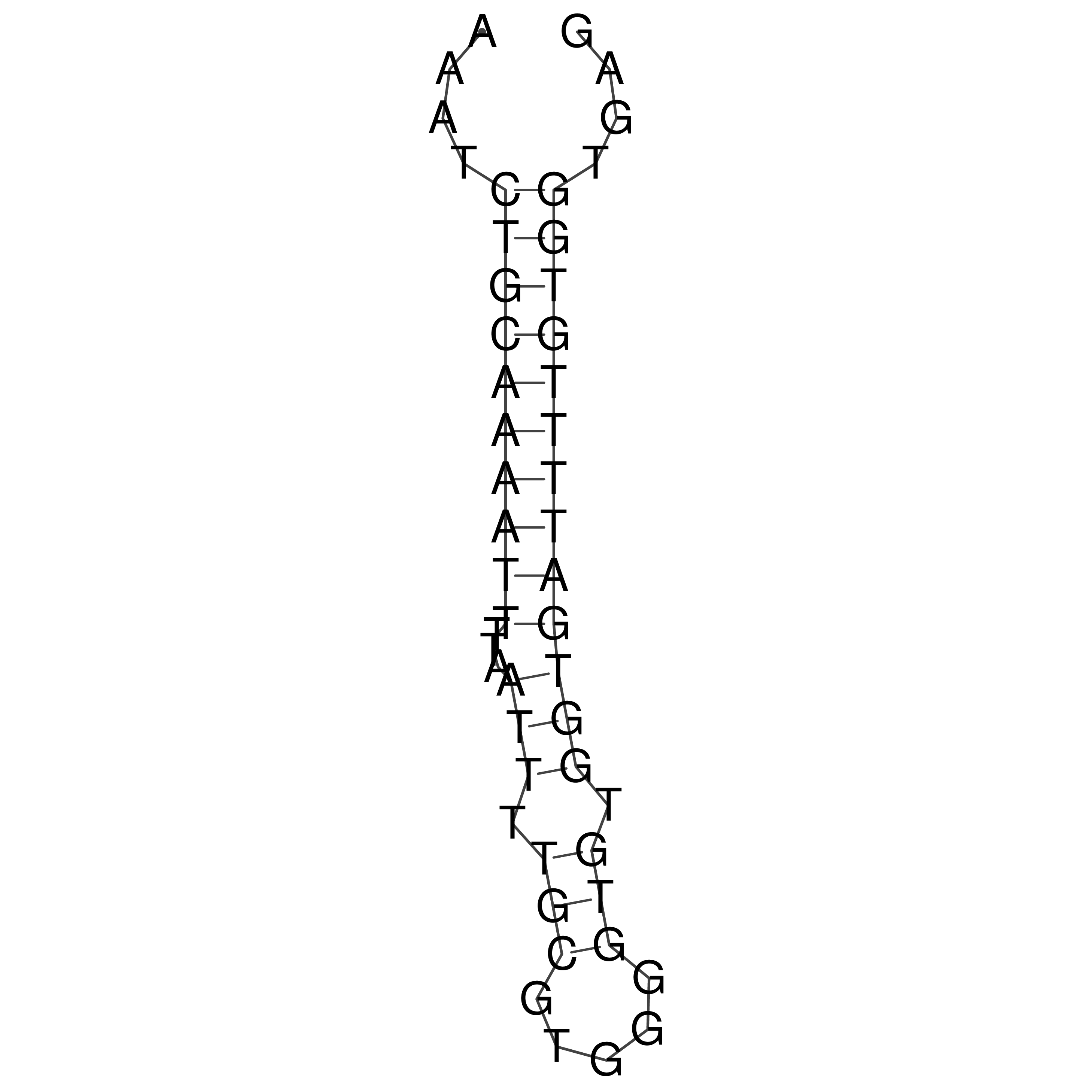
Relaxase
| ID | 2336 | GenBank | WP_004153015 |
| Name | traI_HTH42_RS00610_pG12-KPC-2 |
UniProt ID | A0A8A5TN24 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190492.51 Da Isoelectric Point: 6.3661
>WP_004153015.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGMFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRHNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGASGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQKSVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTSFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNAPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTQAQEKAIQ
KALSEGETLPAGQPASLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGMFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRHNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGASGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQKSVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTSFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNAPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTQAQEKAIQ
KALSEGETLPAGQPASLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2064 | GenBank | WP_004152620 |
| Name | traD_HTH42_RS00605_pG12-KPC-2 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85871.84 Da Isoelectric Point: 5.0101
>WP_004152620.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDTPEPGPADTDSHASEQPEPVSPPA
PAVMTVTPAPVKSPPTTKRPAAEPSVRATEPPVLRGTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDTPEPGPADTDSHASEQPEPVSPPA
PAVMTVTPAPVKSPPTTKRPAAEPSVRATEPPVLRGTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 72384..101381
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTH42_RS00395 (pG12-KPC-2_40) | 67486..67836 | + | 351 | WP_004153414 | hypothetical protein | - |
| HTH42_RS00400 (pG12-KPC-2_42) | 68466..68819 | + | 354 | WP_004152748 | hypothetical protein | - |
| HTH42_RS00405 (pG12-KPC-2_43) | 68876..69223 | + | 348 | WP_004152749 | hypothetical protein | - |
| HTH42_RS00410 | 69318..69464 | + | 147 | WP_004152750 | hypothetical protein | - |
| HTH42_RS00415 (pG12-KPC-2_44) | 69515..70348 | + | 834 | WP_004152751 | N-6 DNA methylase | - |
| HTH42_RS00420 (pG12-KPC-2_46) | 71168..71989 | + | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| HTH42_RS00425 (pG12-KPC-2_47) | 72022..72351 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| HTH42_RS00430 (pG12-KPC-2_48) | 72384..72869 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| HTH42_RS00435 | 73291..73689 | + | 399 | WP_004152493 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HTH42_RS00440 | 73863..74549 | + | 687 | WP_004152494 | transcriptional regulator TraJ family protein | - |
| HTH42_RS00445 | 74628..75014 | + | 387 | WP_004152495 | TraY domain-containing protein | - |
| HTH42_RS00450 | 75068..75436 | + | 369 | WP_004152496 | type IV conjugative transfer system pilin TraA | - |
| HTH42_RS00455 | 75450..75755 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| HTH42_RS00460 | 75775..76341 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| HTH42_RS00465 | 76328..77068 | + | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| HTH42_RS00470 | 77068..78492 | + | 1425 | WP_004155033 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HTH42_RS00475 | 78606..79190 | + | 585 | WP_004161368 | type IV conjugative transfer system lipoprotein TraV | traV |
| HTH42_RS00480 (pG12-KPC-2_49) | 79322..79732 | + | 411 | WP_004152499 | hypothetical protein | - |
| HTH42_RS00485 | 79838..80056 | + | 219 | WP_004152501 | hypothetical protein | - |
| HTH42_RS00490 (pG12-KPC-2_50) | 80057..80368 | + | 312 | WP_004152502 | hypothetical protein | - |
| HTH42_RS00495 (pG12-KPC-2_51) | 80435..80839 | + | 405 | WP_004152503 | hypothetical protein | - |
| HTH42_RS00500 (pG12-KPC-2_52) | 80882..81271 | + | 390 | WP_004153076 | hypothetical protein | - |
| HTH42_RS00505 (pG12-KPC-2_53) | 81279..81677 | + | 399 | WP_011977783 | hypothetical protein | - |
| HTH42_RS00510 | 81749..84388 | + | 2640 | WP_004152505 | type IV secretion system protein TraC | virb4 |
| HTH42_RS00515 | 84388..84777 | + | 390 | WP_004152506 | type-F conjugative transfer system protein TrbI | - |
| HTH42_RS00520 | 84777..85403 | + | 627 | WP_004152507 | type-F conjugative transfer system protein TraW | traW |
| HTH42_RS00525 (pG12-KPC-2_54) | 85445..85834 | + | 390 | WP_004152508 | hypothetical protein | - |
| HTH42_RS00530 | 85831..86820 | + | 990 | WP_011977785 | conjugal transfer pilus assembly protein TraU | traU |
| HTH42_RS00535 | 86833..87471 | + | 639 | WP_004152672 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HTH42_RS00540 | 87530..89485 | + | 1956 | WP_004152673 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HTH42_RS00545 | 89517..89771 | + | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| HTH42_RS00550 (pG12-KPC-2_55) | 89749..89997 | + | 249 | WP_004152675 | hypothetical protein | - |
| HTH42_RS00555 (pG12-KPC-2_56) | 90010..90336 | + | 327 | WP_004152676 | hypothetical protein | - |
| HTH42_RS00560 | 90357..91109 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HTH42_RS00565 | 91120..91359 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HTH42_RS00570 | 91331..91888 | + | 558 | WP_004152678 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HTH42_RS00575 | 91934..92377 | + | 444 | WP_004152679 | F-type conjugal transfer protein TrbF | - |
| HTH42_RS00580 | 92355..93734 | + | 1380 | WP_004165169 | conjugal transfer pilus assembly protein TraH | traH |
| HTH42_RS00585 | 93734..96583 | + | 2850 | WP_072241631 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HTH42_RS00590 | 96596..97144 | + | 549 | WP_004152623 | conjugal transfer entry exclusion protein TraS | - |
| HTH42_RS00595 | 97328..98059 | + | 732 | WP_004152622 | conjugal transfer complement resistance protein TraT | - |
| HTH42_RS00600 (pG12-KPC-2_57) | 98251..98940 | + | 690 | WP_004198206 | hypothetical protein | - |
| HTH42_RS00605 | 99069..101381 | + | 2313 | WP_004152620 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 3559 | GenBank | NZ_KU665642 |
| Plasmid name | pG12-KPC-2 | Incompatibility group | IncFIB |
| Plasmid size | 111926 bp | Coordinate of oriT [Strand] | 72942..72991 [-] |
| Host baterium | Klebsiella pneumoniae strain K47-25 |
Cargo genes
| Drug resistance gene | blaKPC-2, blaOXA-9 |
| Virulence gene | - |
| Metal resistance gene | merD, merA |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |