Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103102 |
| Name | oriT_p80-547 |
| Organism | Shigella dysenteriae strain 80-547 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KT754160 (68008..68131 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 101..106, 119..124 (TTTAAT..ATTAAA) 91..99, 113..121 (AATAATGTA..TACATTATT) 90..95, 107..112 (AAATAA..TTATTT) 57..62, 70..75 (TGATTT..AAATCA) 41..48, 61..68 (AAAAACAA..TTGTTTTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGTTTTTTAAATCATTAGTTTATGTTCTAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file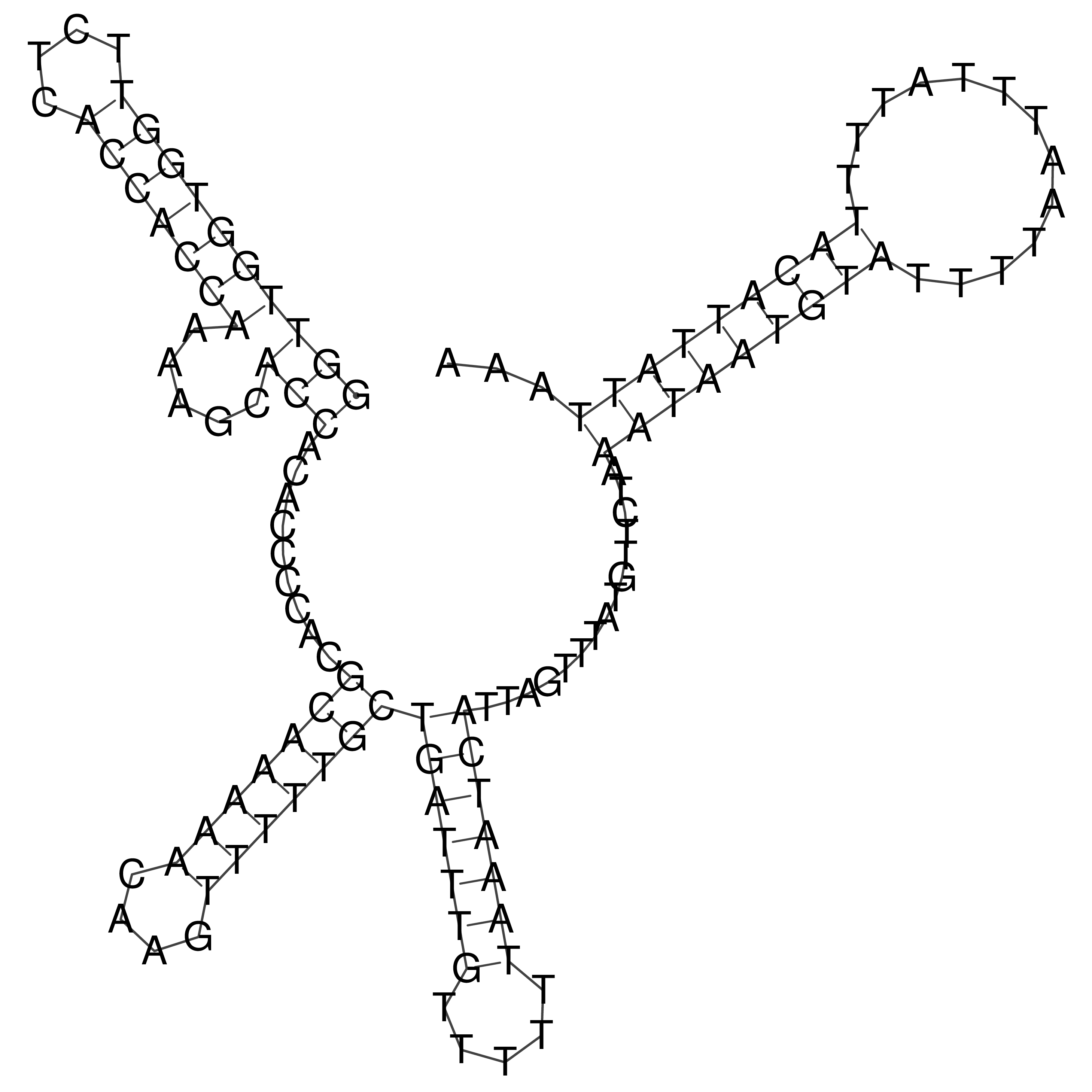
Relaxase
| ID | 2324 | GenBank | WP_171265027 |
| Name | traI_HPF59_RS00555_p80-547 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192245.06 Da Isoelectric Point: 6.0277
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNRHRPGYDLTFSAPKSASMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLPEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAVSVKAGEESVAQVSGVREQAMLTQTIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVFGGDRLQVSSVSEDAMTVVVPGRAEPATLPVSDSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KTRAGETSLETAISHQKSALHTPAQQAIHLALPVVESKNLAFSMVDLLTEAKSFAAEGTGFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVMPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMHSAGVDAQTLASFLHDT
QLQQRSGEIPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTEQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVNNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRLGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRMIRPAQERVEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVF
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVMAEVQAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVREIAGQERDRTAITEREAALPESVLREPQRVREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 984 | GenBank | WP_001541914 |
| Name | WP_001541914_p80-547 |
UniProt ID | A0A142CLH0 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14544.56 Da Isoelectric Point: 5.0359
MARVNLYISNEVHEKINMIVEKRRQEGARDKDISLSGTASMLLELGLRVYDAQMERKESAFNQTEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGNPKFEYASMVDDIREKVSSEMERFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A142CLH0 |
| ID | 985 | GenBank | WP_001254386 |
| Name | WP_001254386_p80-547 |
UniProt ID | A0A3Z6KJB5 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9005.11 Da Isoelectric Point: 10.1422
MRRRNARGGISRTVSVYLDEDTNNRLIKAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVTEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3Z6KJB5 |
T4CP
| ID | 2048 | GenBank | WP_001064245 |
| Name | traC_HPF59_RS00470_p80-547 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99159.81 Da Isoelectric Point: 5.8486
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASITTQTVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNVADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQYT
ANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2049 | GenBank | WP_000009363 |
| Name | traD_HPF59_RS00550_p80-547 |
UniProt ID | _ |
| Length | 741 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 741 a.a. Molecular weight: 84259.95 Da Isoelectric Point: 5.0504
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMA
AYEAWQQENHPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 67436..93086
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HPF59_RS00760 | 62815..63591 | + | 777 | WP_201799183 | hypothetical protein | - |
| HPF59_RS00365 | 64005..64430 | + | 426 | WP_001198918 | antirestriction protein | - |
| HPF59_RS00370 | 64477..64899 | + | 423 | WP_000271685 | DUF1380 family protein | - |
| HPF59_RS00375 | 64896..65083 | + | 188 | Protein_77 | hypothetical protein | - |
| HPF59_RS00380 | 65053..65610 | - | 558 | WP_201799181 | hypothetical protein | - |
| HPF59_RS00390 | 65913..66200 | + | 288 | WP_171265023 | hypothetical protein | - |
| HPF59_RS00395 | 66319..67140 | + | 822 | Protein_80 | DUF932 domain-containing protein | - |
| HPF59_RS00400 | 67436..68038 | - | 603 | WP_326305067 | transglycosylase SLT domain-containing protein | - |
| HPF59_RS00405 | 68360..68743 | + | 384 | WP_001541914 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HPF59_RS00410 | 68928..69614 | + | 687 | WP_000332492 | PAS domain-containing protein | - |
| HPF59_RS00415 | 69708..69935 | + | 228 | WP_001254386 | conjugal transfer relaxosome protein TraY | - |
| HPF59_RS00420 | 69969..70334 | + | 366 | WP_000340270 | type IV conjugative transfer system pilin TraA | - |
| HPF59_RS00425 | 70349..70660 | + | 312 | WP_064221663 | type IV conjugative transfer system protein TraL | traL |
| HPF59_RS00430 | 70682..71248 | + | 567 | WP_000399797 | type IV conjugative transfer system protein TraE | traE |
| HPF59_RS00435 | 71235..71963 | + | 729 | WP_001230766 | type-F conjugative transfer system secretin TraK | traK |
| HPF59_RS00440 | 71963..73390 | + | 1428 | WP_000146655 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HPF59_RS00445 | 73380..73970 | + | 591 | WP_001523507 | conjugal transfer pilus-stabilizing protein TraP | - |
| HPF59_RS00450 | 73957..74154 | + | 198 | WP_001454499 | conjugal transfer protein TrbD | - |
| HPF59_RS00455 | 74166..74417 | + | 252 | WP_001038342 | conjugal transfer protein TrbG | - |
| HPF59_RS00460 | 74414..74929 | + | 516 | WP_171265024 | type IV conjugative transfer system lipoprotein TraV | traV |
| HPF59_RS00465 | 75064..75285 | + | 222 | WP_001278689 | conjugal transfer protein TraR | - |
| HPF59_RS00470 | 75445..78072 | + | 2628 | WP_001064245 | type IV secretion system protein TraC | virb4 |
| HPF59_RS00475 | 78069..78455 | + | 387 | WP_000099689 | type-F conjugative transfer system protein TrbI | - |
| HPF59_RS00480 | 78452..79084 | + | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| HPF59_RS00485 | 79081..80073 | + | 993 | WP_000830839 | conjugal transfer pilus assembly protein TraU | traU |
| HPF59_RS00490 | 80082..80720 | + | 639 | WP_171265032 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HPF59_RS00495 | 80717..82525 | + | 1809 | WP_171265025 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HPF59_RS00500 | 82552..82809 | + | 258 | WP_000864325 | conjugal transfer protein TrbE | - |
| HPF59_RS00505 | 82802..83545 | + | 744 | WP_001030376 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HPF59_RS00510 | 83561..83908 | + | 348 | WP_025269861 | conjugal transfer protein TrbA | - |
| HPF59_RS00515 | 84027..84311 | + | 285 | WP_000624110 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HPF59_RS00520 | 84298..84843 | + | 546 | WP_171265026 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HPF59_RS00525 | 84773..85135 | + | 363 | WP_001443292 | P-type conjugative transfer protein TrbJ | - |
| HPF59_RS00530 | 85135..86508 | + | 1374 | WP_001137358 | conjugal transfer pilus assembly protein TraH | traH |
| HPF59_RS00535 | 86505..89327 | + | 2823 | WP_001007026 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HPF59_RS00540 | 89342..89845 | + | 504 | WP_061583830 | surface exclusion protein | - |
| HPF59_RS00545 | 89877..90608 | + | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| HPF59_RS00550 | 90861..93086 | + | 2226 | WP_000009363 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 3545 | GenBank | NZ_KT754160 |
| Plasmid name | p80-547 | Incompatibility group | IncFIB |
| Plasmid size | 129841 bp | Coordinate of oriT [Strand] | 68008..68131 [+] |
| Host baterium | Shigella dysenteriae strain 80-547 |
Cargo genes
| Drug resistance gene | blaTEM-1B, sul1, qacE, ant(3'')-Ia, catA1, tet(B) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |