Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103085 |
| Name | oriT_pEC14III |
| Organism | Escherichia coli |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KU932028 (6851..6974 [-], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 92..99, 113..120 (ATAATGTA..TACATTAT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTATTTGAATCATTAACTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATAAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file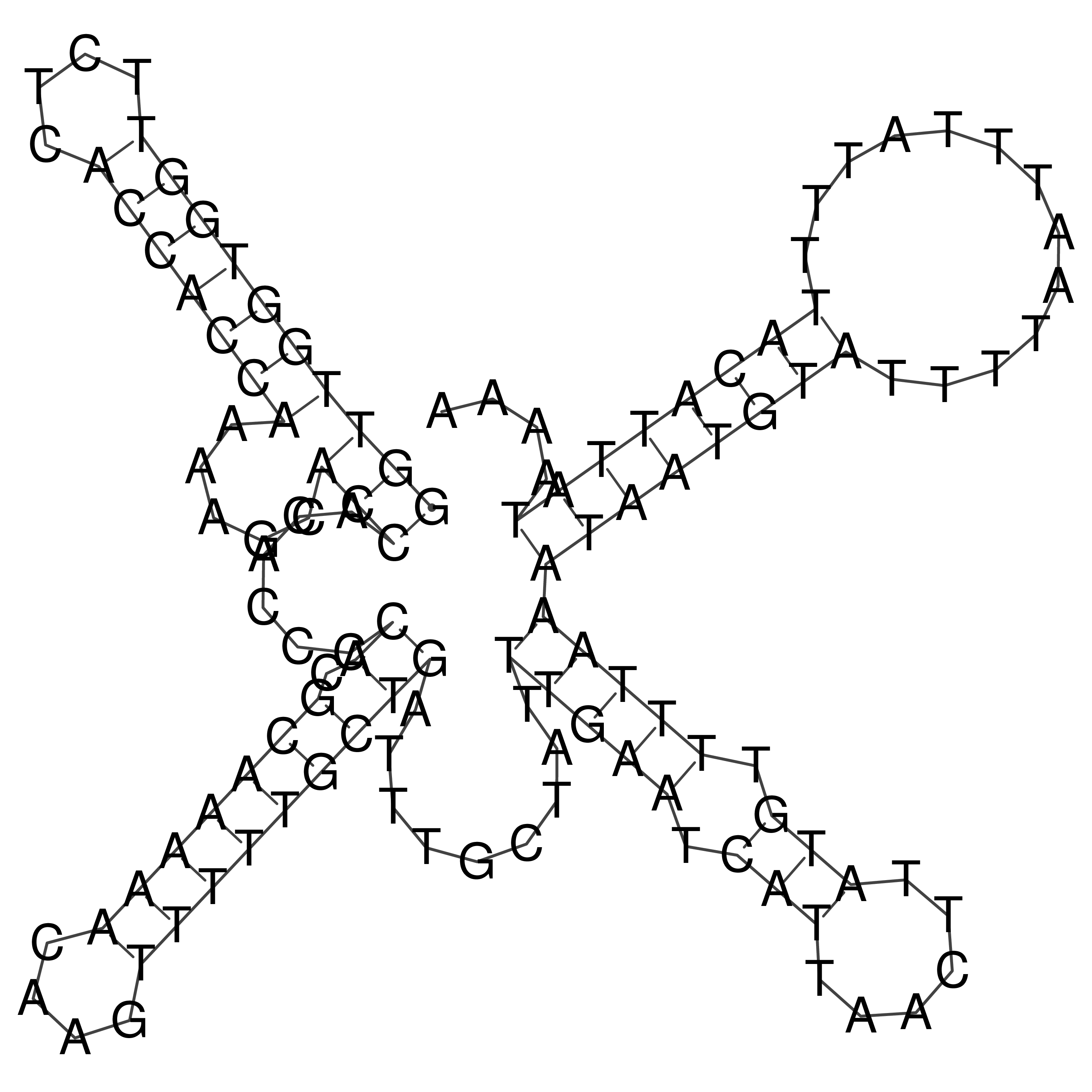
Auxiliary protein
| ID | 970 | GenBank | WP_001254388 |
| Name | WP_001254388_pEC14III |
UniProt ID | W9A0Z2 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9072.20 Da Isoelectric Point: 10.2071
MRRRNARGGISRTVSVYLDEDTNNRLIRAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVIEENES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | W9A0Z2 |
| ID | 971 | GenBank | WP_001151564 |
| Name | WP_001151564_pEC14III |
UniProt ID | A0AA36PD43 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14446.39 Da Isoelectric Point: 4.7197
MAKVQAYVSDEIVYKINKIVERRRAEGAKSTDVSFSSISTMLLELGLRVYEAQMERKESAFNQAEFNKVL
LECAVKTQSTVAKILGIESLSPHVSGNPKFEYANMVEDIRDKVSSEMERFFPENDEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2030 | GenBank | WP_072749386 |
| Name | traD_HTH55_RS00370_pEC14III |
UniProt ID | _ |
| Length | 729 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 729 a.a. Molecular weight: 82874.49 Da Isoelectric Point: 5.0524
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTENPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVISGEDVTQAEQPQQPQQPQQ
PQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQENHPD
IQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2031 | GenBank | WP_064558378 |
| Name | traC_HTH55_RS00470_pEC14III |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 99275.99 Da Isoelectric Point: 6.3025
MSNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAI
PINGANKSIVEALDHMLRTKLPRDIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAA
ATQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASIATQTVDAQAFIDIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAF
LWNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALKVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAGSPTIRSRLDEMIVLLDQY
TANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGW
RLLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILRQSAKEFAKY
NQLYPDQFQPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEG
LSIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 58901..78358
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTH55_RS00370 | 58901..61090 | - | 2190 | WP_072749386 | type IV conjugative transfer system coupling protein TraD | virb4 |
| HTH55_RS00375 | 61141..61881 | - | 741 | WP_021514832 | hypothetical protein | - |
| HTH55_RS00380 | 62084..62815 | - | 732 | WP_000782451 | conjugal transfer complement resistance protein TraT | - |
| HTH55_RS00385 | 62864..63349 | - | 486 | WP_000605868 | hypothetical protein | - |
| HTH55_RS00390 | 63365..66187 | - | 2823 | WP_021532657 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HTH55_RS00395 | 66184..67557 | - | 1374 | WP_001137358 | conjugal transfer pilus assembly protein TraH | traH |
| HTH55_RS00400 | 67557..67919 | - | 363 | WP_010379880 | P-type conjugative transfer protein TrbJ | - |
| HTH55_RS00405 | 67849..68394 | - | 546 | WP_000052618 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HTH55_RS00410 | 68381..68617 | - | 237 | WP_001346259 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HTH55_RS00415 | 68778..69521 | - | 744 | WP_001030376 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HTH55_RS00420 | 69514..69771 | - | 258 | WP_000864341 | conjugal transfer protein TrbE | - |
| HTH55_RS00425 | 69798..71648 | - | 1851 | WP_021537336 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HTH55_RS00430 | 71645..72067 | - | 423 | WP_001229306 | HNH endonuclease signature motif containing protein | - |
| HTH55_RS00435 | 72092..72463 | - | 372 | WP_021514849 | hypothetical protein | - |
| HTH55_RS00440 | 72460..73098 | - | 639 | WP_021514850 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HTH55_RS00445 | 73339..73530 | + | 192 | WP_032147853 | hypothetical protein | - |
| HTH55_RS00450 | 73527..73706 | - | 180 | WP_032193743 | hypothetical protein | - |
| HTH55_RS00455 | 73727..74719 | - | 993 | WP_000830838 | conjugal transfer pilus assembly protein TraU | traU |
| HTH55_RS00460 | 74716..75348 | - | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| HTH55_RS00465 | 75345..75731 | - | 387 | WP_021536235 | type-F conjugative transfer system protein TrbI | - |
| HTH55_RS00470 | 75728..78358 | - | 2631 | WP_064558378 | type IV secretion system protein TraC | virb4 |
| HTH55_RS00475 | 78484..78846 | - | 363 | WP_021514859 | hypothetical protein | - |
| HTH55_RS00480 | 78874..79089 | - | 216 | WP_000556747 | hypothetical protein | - |
| HTH55_RS00485 | 79169..79645 | - | 477 | WP_000549580 | hypothetical protein | - |
| HTH55_RS00490 | 79638..79859 | - | 222 | WP_001278695 | conjugal transfer protein TraR | - |
Host bacterium
| ID | 3528 | GenBank | NZ_KU932028 |
| Plasmid name | pEC14III | Incompatibility group | IncFII |
| Plasmid size | 80057 bp | Coordinate of oriT [Strand] | 6851..6974 [-] |
| Host baterium | Escherichia coli |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |