Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103062 |
| Name | oriT_pYD786-2 |
| Organism | Escherichia coli strain YD786 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KU254579 (37168..37291 [-], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 101..106, 119..124 (TTTAAT..ATTAAA) 91..99, 113..121 (AATAATGTA..TACATTATT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTTTTTGAATCATTAGCTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file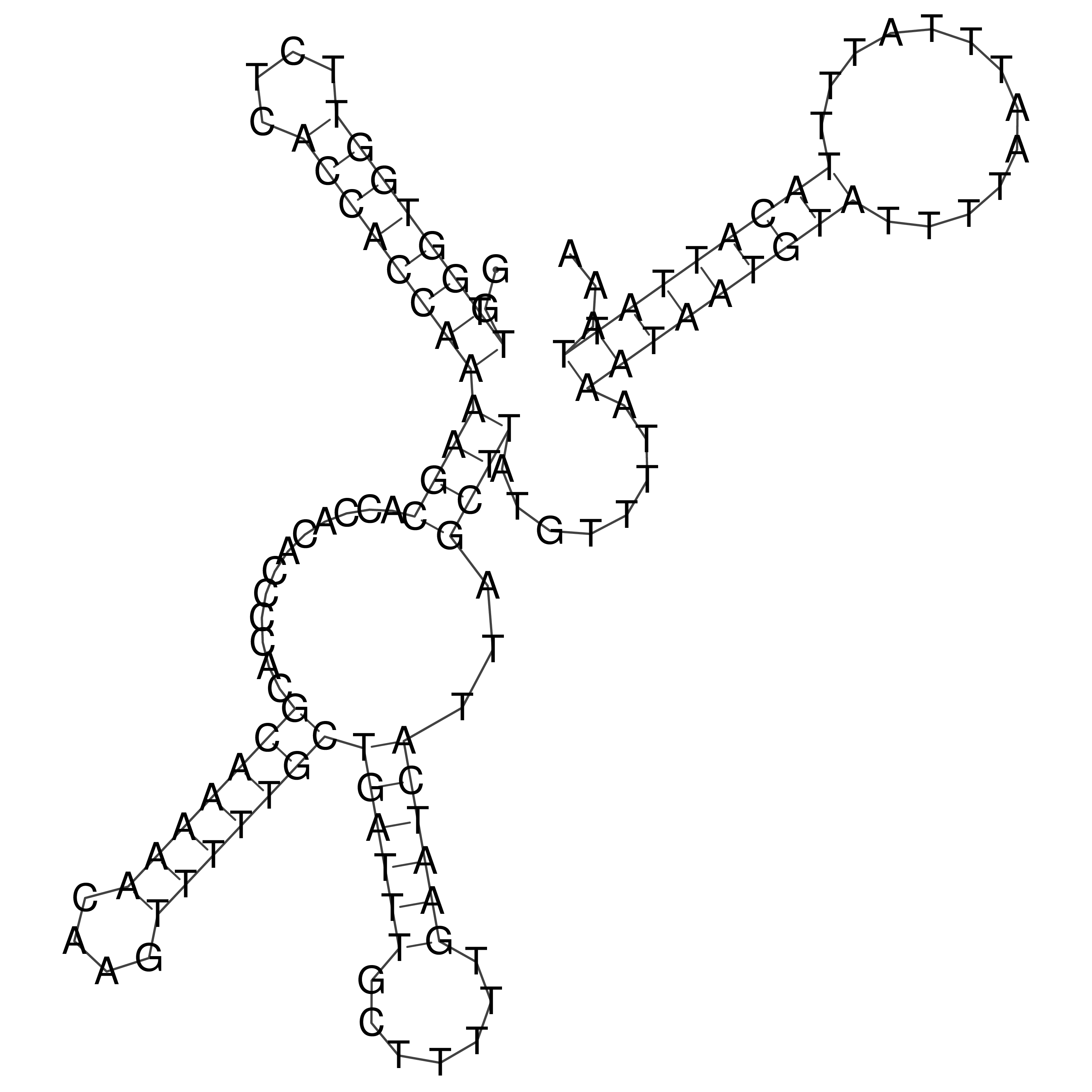
Relaxase
| ID | 2292 | GenBank | WP_103687900 |
| Name | traI_HTG80_RS00045_pYD786-2 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192060.79 Da Isoelectric Point: 5.8861
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRLATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVTSVSEDAMTVVVPGRAEPASLPVSDSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGEISLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTSFVDLGREIDA
QIKRGDLLHVDVAKGYGTDLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQESAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVF
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTSDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTELAVREIAGQERDRAAITEREAALPESVLREPQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 952 | GenBank | WP_001254386 |
| Name | WP_001254386_pYD786-2 |
UniProt ID | A0A3Z6KJB5 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9005.11 Da Isoelectric Point: 10.1422
MRRRNARGGISRTVSVYLDEDTNNRLIKAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVTEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3Z6KJB5 |
| ID | 953 | GenBank | WP_000124981 |
| Name | WP_000124981_pYD786-2 |
UniProt ID | A0A630FTW9 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14542.59 Da Isoelectric Point: 5.0278
MARVNLYISNEVHEKINMIVEKRRQEGARDKDISLSGTASMLLELGLRVYDAQMERKESAFNQTEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGNPKFEYASMVDDIREKVSVEMDRFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A630FTW9 |
T4CP
| ID | 2000 | GenBank | WP_130989118 |
| Name | traD_HTG80_RS00050_pYD786-2 |
UniProt ID | _ |
| Length | 740 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 740 a.a. Molecular weight: 84076.74 Da Isoelectric Point: 5.0504
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTSGKEDPFW
QGSGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTYLRNSPAANLVEEKIEKTAISIRAVLTNYV
KAIRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNR
RVWFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHK
IAEFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKL
SLKYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQP
QQPQQPQQPQQPQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAA
YEAWQQENHPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2001 | GenBank | WP_000069764 |
| Name | traC_HTG80_RS00130_pYD786-2 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 99302.93 Da Isoelectric Point: 5.8486
MSNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAI
PINGANESIVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMNAA
ATQFPLPEGMNLPLTLRHYRVFFSYCSPSKKKSRADILEMENLVKIIRASLQGASITTQAVDAQAFIDIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLREKGRNSTARILNFHLARNPEIAF
LWNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQY
TANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGW
RLLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKY
NQLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEG
LSIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 11273..37863
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTG80_RS00050 | 11273..13495 | - | 2223 | WP_130989118 | type IV conjugative transfer system coupling protein TraD | virb4 |
| HTG80_RS00055 | 13747..14478 | - | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| HTG80_RS00060 | 14492..15001 | - | 510 | WP_000628099 | conjugal transfer entry exclusion protein TraS | - |
| HTG80_RS00065 | 14998..17823 | - | 2826 | WP_001007030 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HTG80_RS00070 | 17820..19193 | - | 1374 | WP_001137358 | conjugal transfer pilus assembly protein TraH | traH |
| HTG80_RS00075 | 19193..19555 | - | 363 | WP_010379880 | P-type conjugative transfer protein TrbJ | - |
| HTG80_RS00080 | 19485..20030 | - | 546 | WP_000060037 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HTG80_RS00085 | 20017..20301 | - | 285 | WP_000624108 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HTG80_RS00090 | 20420..20767 | - | 348 | WP_001287893 | conjugal transfer protein TrbA | - |
| HTG80_RS00095 | 20783..21526 | - | 744 | WP_001030376 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HTG80_RS00100 | 21519..21776 | - | 258 | WP_000864325 | conjugal transfer protein TrbE | - |
| HTG80_RS00105 | 21803..23611 | - | 1809 | WP_001567342 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HTG80_RS00110 | 23608..24246 | - | 639 | WP_029400171 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HTG80_RS00115 | 24255..25247 | - | 993 | WP_000830839 | conjugal transfer pilus assembly protein TraU | traU |
| HTG80_RS00120 | 25244..25876 | - | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| HTG80_RS00125 | 25873..26259 | - | 387 | WP_029400173 | type-F conjugative transfer system protein TrbI | - |
| HTG80_RS00130 | 26256..28886 | - | 2631 | WP_000069764 | type IV secretion system protein TraC | virb4 |
| HTG80_RS00135 | 29013..29360 | - | 348 | WP_000802783 | hypothetical protein | - |
| HTG80_RS00140 | 29388..29605 | - | 218 | Protein_28 | hypothetical protein | - |
| HTG80_RS00145 | 29685..30161 | - | 477 | WP_000549579 | hypothetical protein | - |
| HTG80_RS00150 | 30154..30375 | - | 222 | WP_001278695 | conjugal transfer protein TraR | - |
| HTG80_RS00155 | 30510..31025 | - | 516 | WP_000809874 | type IV conjugative transfer system lipoprotein TraV | traV |
| HTG80_RS00160 | 31022..31342 | - | 321 | WP_001057307 | conjugal transfer protein TrbD | - |
| HTG80_RS00165 | 31329..31919 | - | 591 | WP_000002784 | conjugal transfer pilus-stabilizing protein TraP | - |
| HTG80_RS00170 | 31909..33336 | - | 1428 | WP_000146657 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HTG80_RS00175 | 33336..34064 | - | 729 | WP_001230798 | type-F conjugative transfer system secretin TraK | traK |
| HTG80_RS00180 | 34051..34617 | - | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| HTG80_RS00185 | 34639..34950 | - | 312 | WP_000012103 | type IV conjugative transfer system protein TraL | traL |
| HTG80_RS00190 | 34965..35324 | - | 360 | WP_000340272 | type IV conjugative transfer system pilin TraA | - |
| HTG80_RS00195 | 35358..35585 | - | 228 | WP_001254386 | conjugal transfer relaxosome protein TraY | - |
| HTG80_RS00200 | 35679..36365 | - | 687 | WP_000332484 | PAS domain-containing protein | - |
| HTG80_RS00205 | 36556..36939 | - | 384 | WP_000124981 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HTG80_RS00210 | 37216..37863 | + | 648 | WP_000614934 | transglycosylase SLT domain-containing protein | virB1 |
| HTG80_RS00215 | 38160..38981 | - | 822 | WP_021573168 | DUF932 domain-containing protein | - |
| HTG80_RS00220 | 39098..39385 | - | 288 | WP_000107538 | hypothetical protein | - |
| HTG80_RS00225 | 39636..40640 | + | 1005 | WP_024257664 | IS110 family transposase | - |
| HTG80_RS00440 | 41064..41225 | + | 162 | Protein_46 | hypothetical protein | - |
| HTG80_RS00445 | 41235..41465 | - | 231 | WP_001426396 | hypothetical protein | - |
| HTG80_RS00235 | 41685..41810 | - | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| HTG80_RS00450 | 41752..41901 | - | 150 | Protein_49 | plasmid maintenance protein Mok | - |
| HTG80_RS00240 | 42080..42842 | - | 763 | Protein_50 | plasmid SOS inhibition protein A | - |
Host bacterium
| ID | 3505 | GenBank | NZ_KU254579 |
| Plasmid name | pYD786-2 | Incompatibility group | IncFII |
| Plasmid size | 68812 bp | Coordinate of oriT [Strand] | 37168..37291 [-] |
| Host baterium | Escherichia coli strain YD786 |
Cargo genes
| Drug resistance gene | fosA6, floR |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |