Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102980 |
| Name | oriT_pGR-1870 |
| Organism | Klebsiella pneumoniae |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_023905 (80687..80736 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pGR-1870
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTCATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTCATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file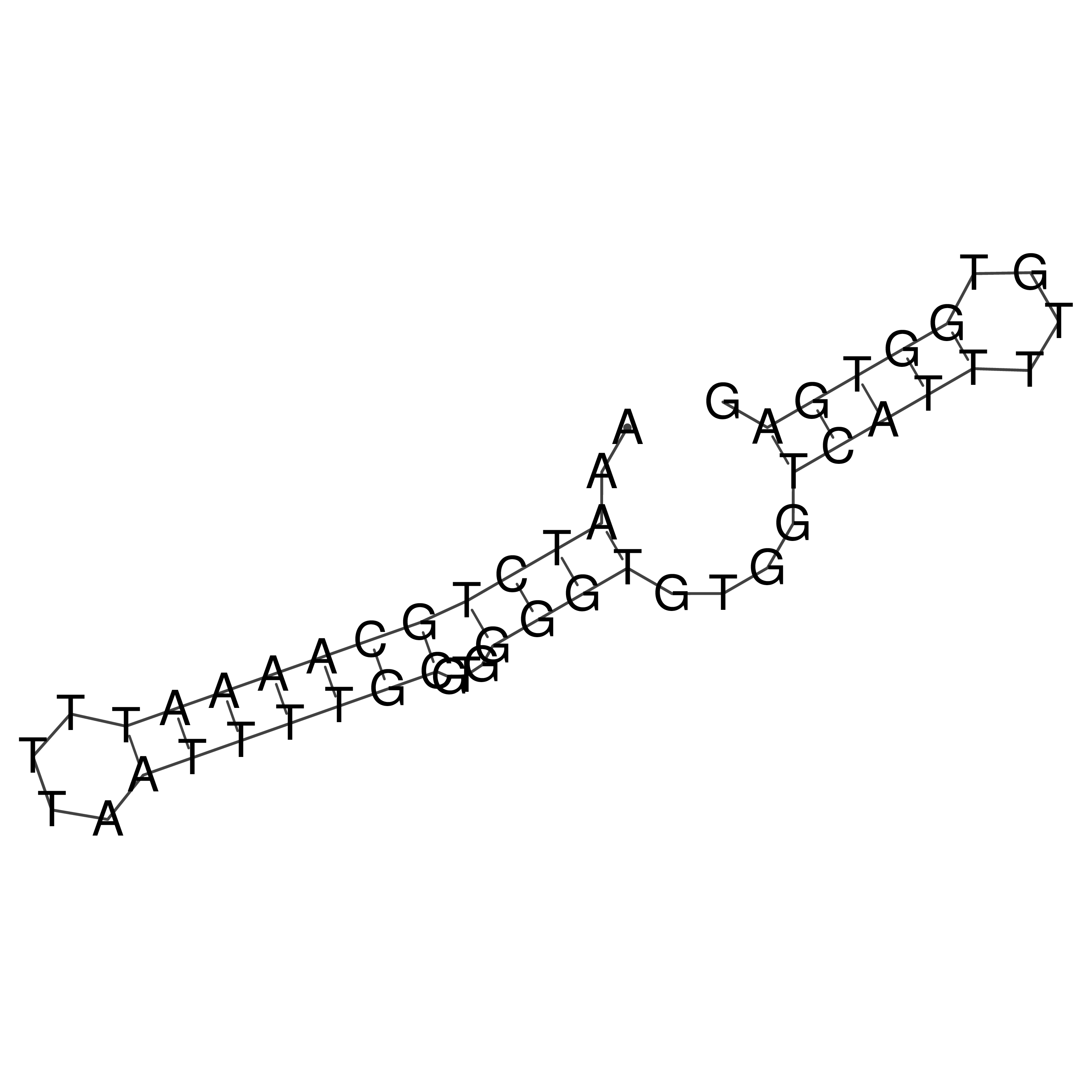
Relaxase
| ID | 2235 | GenBank | WP_024269743 |
| Name | traI_HTC14_RS00685_pGR-1870 |
UniProt ID | W6AUB9 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190506.54 Da Isoelectric Point: 6.3661
>WP_024269743.1 conjugative transfer relaxase/helicase TraI [Klebsiella pneumoniae]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGMFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRHNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGASGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQKSVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTSFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNAPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTQAQEKAIQ
KALSEGETLPAGQPASLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGMFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRHNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGASGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQKSVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTSFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNAPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTQAQEKAIQ
KALSEGETLPAGQPASLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1924 | GenBank | WP_004152620 |
| Name | traD_HTC14_RS00680_pGR-1870 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85871.84 Da Isoelectric Point: 5.0101
>WP_004152620.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDTPEPGPADTDSHASEQPEPVSPPA
PAVMTVTPAPVKSPPTTKRPAAEPSVRATEPPVLRGTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDTPEPGPADTDSHASEQPEPVSPPA
PAVMTVTPAPVKSPPTTKRPAAEPSVRATEPPVLRGTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 80129..109315
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTC14_RS00450 (D647_p45061) | 75146..75496 | + | 351 | WP_004152758 | hypothetical protein | - |
| HTC14_RS00455 | 75644..75820 | + | 177 | WP_159376550 | hypothetical protein | - |
| HTC14_RS00460 (D647_p45062) | 76132..76488 | + | 357 | WP_004152717 | hypothetical protein | - |
| HTC14_RS00465 (D647_p45063) | 76549..76761 | + | 213 | WP_004152718 | hypothetical protein | - |
| HTC14_RS00470 (D647_p45064) | 76772..76996 | + | 225 | WP_004152719 | hypothetical protein | - |
| HTC14_RS00475 (D647_p45065) | 77077..77397 | + | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| HTC14_RS00480 (D647_p45066) | 77387..77665 | + | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| HTC14_RS00485 (D647_p45067) | 77666..78079 | + | 414 | WP_004152722 | helix-turn-helix domain-containing protein | - |
| HTC14_RS00490 | 78432..78608 | + | 177 | WP_159376550 | hypothetical protein | - |
| HTC14_RS00495 (D647_p45068) | 78913..79734 | + | 822 | WP_004178064 | DUF932 domain-containing protein | - |
| HTC14_RS00500 (D647_p45069) | 79767..80096 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| HTC14_RS00505 (D647_p45070) | 80129..80614 | - | 486 | WP_004178063 | transglycosylase SLT domain-containing protein | virB1 |
| HTC14_RS00510 (D647_p45071) | 81048..81440 | + | 393 | WP_004178062 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HTC14_RS00515 (D647_p45072) | 81654..82349 | + | 696 | WP_004178061 | transcriptional regulator TraJ family protein | - |
| HTC14_RS00520 (D647_p45073) | 82433..82804 | + | 372 | WP_004208838 | TraY domain-containing protein | - |
| HTC14_RS00525 (D647_p45074) | 82858..83226 | + | 369 | WP_004178060 | type IV conjugative transfer system pilin TraA | - |
| HTC14_RS00530 (D647_p45075) | 83240..83545 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| HTC14_RS00535 (D647_p45076) | 83565..84131 | + | 567 | WP_004152602 | type IV conjugative transfer system protein TraE | traE |
| HTC14_RS00540 (D647_p45077) | 84118..84858 | + | 741 | WP_024269739 | type-F conjugative transfer system secretin TraK | traK |
| HTC14_RS00545 (D647_p45078) | 84858..86282 | + | 1425 | WP_024269740 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HTC14_RS00550 (D647_p45079) | 86275..86871 | + | 597 | WP_004152599 | conjugal transfer pilus-stabilizing protein TraP | - |
| HTC14_RS00555 (D647_p45080) | 86894..87463 | + | 570 | WP_004152598 | type IV conjugative transfer system lipoprotein TraV | traV |
| HTC14_RS00560 (D647_p45081) | 87595..88005 | + | 411 | WP_004152597 | lipase chaperone | - |
| HTC14_RS00565 (D647_p45082) | 88010..88300 | + | 291 | WP_004152596 | hypothetical protein | - |
| HTC14_RS00570 (D647_p45083) | 88324..88542 | + | 219 | WP_004171484 | hypothetical protein | - |
| HTC14_RS00575 (D647_p45084) | 88543..88860 | + | 318 | WP_004152595 | hypothetical protein | - |
| HTC14_RS00580 (D647_p45085) | 88927..89331 | + | 405 | WP_004152594 | hypothetical protein | - |
| HTC14_RS00585 | 89627..90025 | + | 399 | WP_004153071 | hypothetical protein | - |
| HTC14_RS00590 (D647_p45086) | 90097..92736 | + | 2640 | WP_004152592 | type IV secretion system protein TraC | virb4 |
| HTC14_RS00595 (D647_p45087) | 92736..93125 | + | 390 | WP_004152591 | type-F conjugative transfer system protein TrbI | - |
| HTC14_RS00600 (D647_p45088) | 93125..93751 | + | 627 | WP_004152590 | type-F conjugative transfer system protein TraW | traW |
| HTC14_RS00605 (D647_p45089) | 93795..94754 | + | 960 | WP_015065634 | conjugal transfer pilus assembly protein TraU | traU |
| HTC14_RS00610 (D647_p45090) | 94767..95405 | + | 639 | WP_004152672 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HTC14_RS00615 (D647_p45091) | 95464..97419 | + | 1956 | WP_004152673 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HTC14_RS00620 (D647_p45092) | 97451..97705 | + | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| HTC14_RS00625 | 97683..97931 | + | 249 | WP_004152675 | hypothetical protein | - |
| HTC14_RS00630 (D647_p45093) | 97944..98270 | + | 327 | WP_004152676 | hypothetical protein | - |
| HTC14_RS00635 (D647_p45094) | 98291..99043 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HTC14_RS00640 (D647_p45095) | 99054..99293 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HTC14_RS00645 (D647_p45096) | 99265..99822 | + | 558 | WP_004152678 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HTC14_RS00650 (D647_p45097) | 99868..100311 | + | 444 | WP_004152679 | F-type conjugal transfer protein TrbF | - |
| HTC14_RS00655 (D647_p45098) | 100289..101668 | + | 1380 | WP_004165169 | conjugal transfer pilus assembly protein TraH | traH |
| HTC14_RS00660 (D647_p45099) | 101668..104517 | + | 2850 | WP_024269741 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HTC14_RS00665 (D647_p45100) | 104530..105078 | + | 549 | WP_004152623 | conjugal transfer entry exclusion protein TraS | - |
| HTC14_RS00670 (D647_p45101) | 105262..105993 | + | 732 | WP_004152622 | conjugal transfer complement resistance protein TraT | - |
| HTC14_RS00675 (D647_p45102) | 106185..106874 | + | 690 | WP_004198206 | hypothetical protein | - |
| HTC14_RS00680 (D647_p45103) | 107003..109315 | + | 2313 | WP_004152620 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 3423 | GenBank | NC_023905 |
| Plasmid name | pGR-1870 | Incompatibility group | IncFII |
| Plasmid size | 116047 bp | Coordinate of oriT [Strand] | 80687..80736 [-] |
| Host baterium | Klebsiella pneumoniae |
Cargo genes
| Drug resistance gene | blaKPC-2, blaOXA-9, blaTEM-1A |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |