Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102959 |
| Name | oriT_pHeBE7 |
| Organism | Escherichia coli strain HeB7 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KT002541 (42314..42437 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 101..106, 119..124 (TTTAAT..ATTAAA) 91..99, 113..121 (AATAATGTA..TACATTATT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTTTTTGAATCATTAGCTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file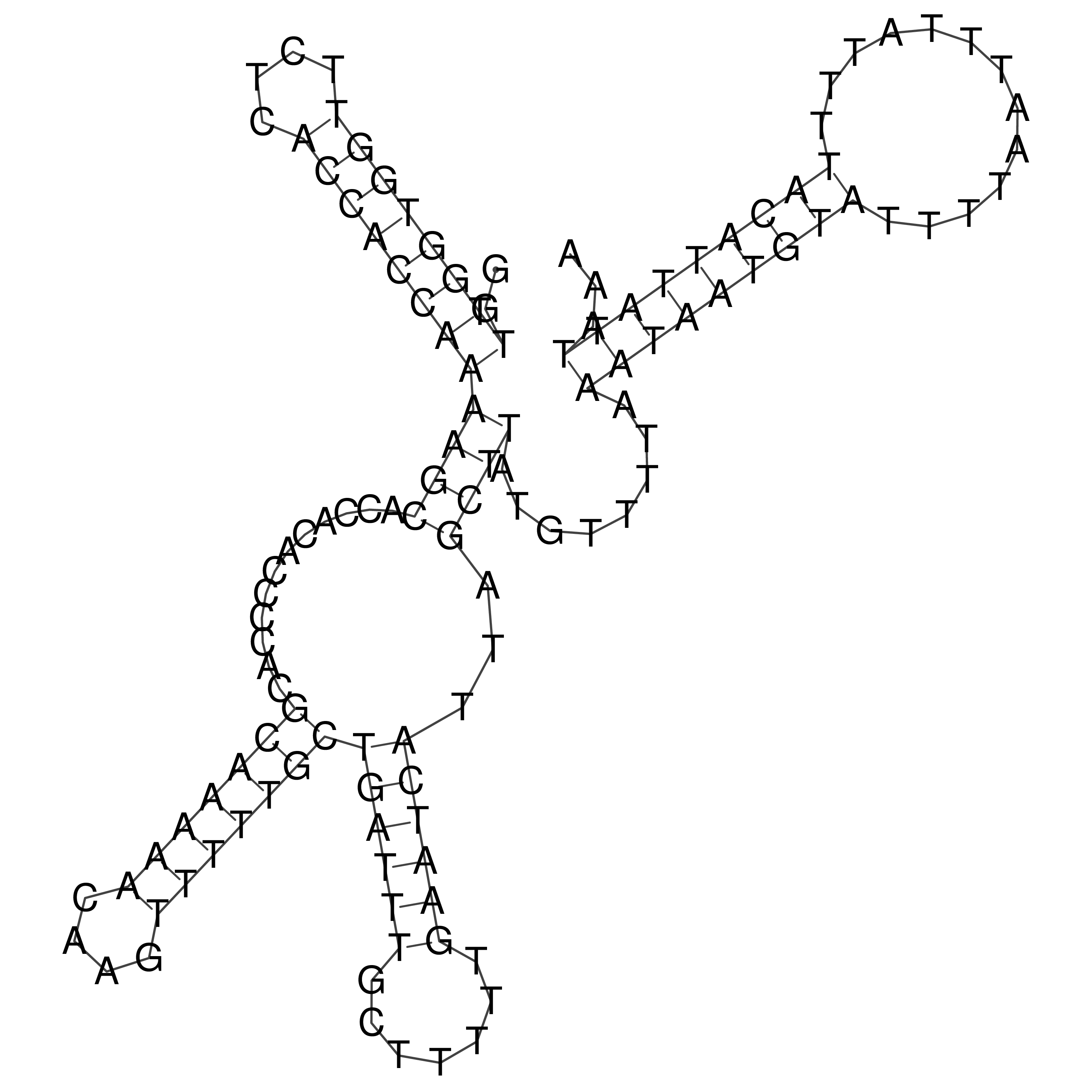
Relaxase
| ID | 2217 | GenBank | WP_015059123 |
| Name | traI_HTF34_RS00405_pHeBE7 |
UniProt ID | I1WZQ7 |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191914.74 Da Isoelectric Point: 6.1060
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMVREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHQEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKISGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFTALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKTGLHTPAQQAIHLALPVVESKNLAFSMVDLLTEAKSFAAEGTSFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLLQRSGETPNFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRLEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMQKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIGLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKSDREVMNAERLFSTARELRDVVAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGTITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVRDIAGLERDRSAISEREAALPESVLREPQRVREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 910 | GenBank | WP_000124981 |
| Name | WP_000124981_pHeBE7 |
UniProt ID | A0A630FTW9 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14542.59 Da Isoelectric Point: 5.0278
MARVNLYISNEVHEKINMIVEKRRQEGARDKDISLSGTASMLLELGLRVYDAQMERKESAFNQTEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGNPKFEYASMVDDIREKVSVEMDRFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A630FTW9 |
| ID | 911 | GenBank | WP_001254386 |
| Name | WP_001254386_pHeBE7 |
UniProt ID | A0A3Z6KJB5 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9005.11 Da Isoelectric Point: 10.1422
MRRRNARGGISRTVSVYLDEDTNNRLIKAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVTEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3Z6KJB5 |
T4CP
| ID | 1904 | GenBank | WP_000069764 |
| Name | traC_HTF34_RS00320_pHeBE7 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 99302.93 Da Isoelectric Point: 5.8486
MSNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAI
PINGANESIVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMNAA
ATQFPLPEGMNLPLTLRHYRVFFSYCSPSKKKSRADILEMENLVKIIRASLQGASITTQAVDAQAFIDIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLREKGRNSTARILNFHLARNPEIAF
LWNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQY
TANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGW
RLLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKY
NQLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEG
LSIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1905 | GenBank | WP_015059124 |
| Name | traD_HTF34_RS00400_pHeBE7 |
UniProt ID | _ |
| Length | 741 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 741 a.a. Molecular weight: 84273.98 Da Isoelectric Point: 5.0524
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTENPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMA
AYEAWQQENHPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 41742..68591
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTF34_RS00205 | 37709..38143 | + | 435 | WP_000845953 | conjugation system SOS inhibitor PsiB | - |
| HTF34_RS00210 | 38140..38902 | + | 763 | Protein_40 | plasmid SOS inhibition protein A | - |
| HTF34_RS00490 | 39081..39230 | + | 150 | Protein_41 | plasmid maintenance protein Mok | - |
| HTF34_RS00215 | 39172..39297 | + | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| HTF34_RS00495 | 39517..39747 | + | 231 | WP_001426396 | hypothetical protein | - |
| HTF34_RS00500 | 39745..39918 | - | 174 | Protein_44 | hypothetical protein | - |
| HTF34_RS00505 | 39988..40194 | + | 207 | WP_000547968 | hypothetical protein | - |
| HTF34_RS00225 | 40219..40506 | + | 288 | WP_000107544 | hypothetical protein | - |
| HTF34_RS00230 | 40624..41445 | + | 822 | WP_001234469 | DUF932 domain-containing protein | - |
| HTF34_RS00235 | 41742..42389 | - | 648 | WP_031943493 | transglycosylase SLT domain-containing protein | virB1 |
| HTF34_RS00240 | 42666..43049 | + | 384 | WP_000124981 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HTF34_RS00245 | 43240..43926 | + | 687 | WP_000332484 | PAS domain-containing protein | - |
| HTF34_RS00250 | 44020..44247 | + | 228 | WP_001254386 | conjugal transfer relaxosome protein TraY | - |
| HTF34_RS00255 | 44281..44640 | + | 360 | WP_000340272 | type IV conjugative transfer system pilin TraA | - |
| HTF34_RS00260 | 44655..44966 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| HTF34_RS00265 | 44988..45554 | + | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| HTF34_RS00270 | 45541..46269 | + | 729 | WP_001230813 | type-F conjugative transfer system secretin TraK | traK |
| HTF34_RS00275 | 46269..47696 | + | 1428 | WP_000146685 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HTF34_RS00280 | 47686..48276 | + | 591 | WP_000002787 | conjugal transfer pilus-stabilizing protein TraP | - |
| HTF34_RS00285 | 48263..48583 | + | 321 | WP_001057287 | conjugal transfer protein TrbD | virb4 |
| HTF34_RS00290 | 48576..48827 | + | 252 | WP_001038342 | conjugal transfer protein TrbG | - |
| HTF34_RS00295 | 48824..49339 | + | 516 | WP_015059130 | type IV conjugative transfer system lipoprotein TraV | traV |
| HTF34_RS00300 | 49474..49695 | + | 222 | WP_001278695 | conjugal transfer protein TraR | - |
| HTF34_RS00305 | 49688..50164 | + | 477 | WP_015059129 | hypothetical protein | - |
| HTF34_RS00310 | 50244..50458 | + | 215 | Protein_63 | hypothetical protein | - |
| HTF34_RS00315 | 50486..50848 | + | 363 | WP_000836675 | hypothetical protein | - |
| HTF34_RS00320 | 50974..53604 | + | 2631 | WP_000069764 | type IV secretion system protein TraC | virb4 |
| HTF34_RS00325 | 53601..53987 | + | 387 | WP_000214092 | type-F conjugative transfer system protein TrbI | - |
| HTF34_RS00330 | 53984..54616 | + | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| HTF34_RS00335 | 54613..55605 | + | 993 | WP_000830839 | conjugal transfer pilus assembly protein TraU | traU |
| HTF34_RS00340 | 55614..56252 | + | 639 | WP_000777692 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HTF34_RS00345 | 56249..58057 | + | 1809 | WP_000821821 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HTF34_RS00350 | 58084..58341 | + | 258 | WP_000864352 | conjugal transfer protein TrbE | - |
| HTF34_RS00355 | 58334..59077 | + | 744 | WP_159122582 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HTF34_RS00360 | 59093..59440 | + | 348 | WP_001287891 | conjugal transfer protein TrbA | - |
| HTF34_RS00365 | 59559..59843 | + | 285 | WP_000624109 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HTF34_RS00370 | 59830..60375 | + | 546 | WP_000059821 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HTF34_RS00375 | 60305..60667 | + | 363 | WP_010379880 | P-type conjugative transfer protein TrbJ | - |
| HTF34_RS00380 | 60667..62040 | + | 1374 | WP_001137358 | conjugal transfer pilus assembly protein TraH | traH |
| HTF34_RS00385 | 62037..64862 | + | 2826 | WP_001007045 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HTF34_RS00390 | 64859..65368 | + | 510 | WP_031943494 | conjugal transfer entry exclusion protein TraS | - |
| HTF34_RS00395 | 65382..66113 | + | 732 | WP_094658456 | conjugal transfer complement resistance protein TraT | - |
| HTF34_RS00400 | 66366..68591 | + | 2226 | WP_015059124 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 3402 | GenBank | NZ_KT002541 |
| Plasmid name | pHeBE7 | Incompatibility group | IncFII |
| Plasmid size | 86015 bp | Coordinate of oriT [Strand] | 42314..42437 [+] |
| Host baterium | Escherichia coli strain HeB7 |
Cargo genes
| Drug resistance gene | blaTEM-1B, rmtB, blaCTX-M-98 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIF11 |