Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102952 |
| Name | oriT_P39|unnamed |
| Organism | Enterococcus faecium isolate P39 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KP345886 (2412..2615 [+], 204 nt) |
| oriT length | 204 nt |
| IRs (inverted repeats) | 125..130, 143..148 (AAATCC..GGATTT) 66..72, 83..89 (ACCCCCC..GGGGGGT) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 204 nt
>oriT_P39|unnamed
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file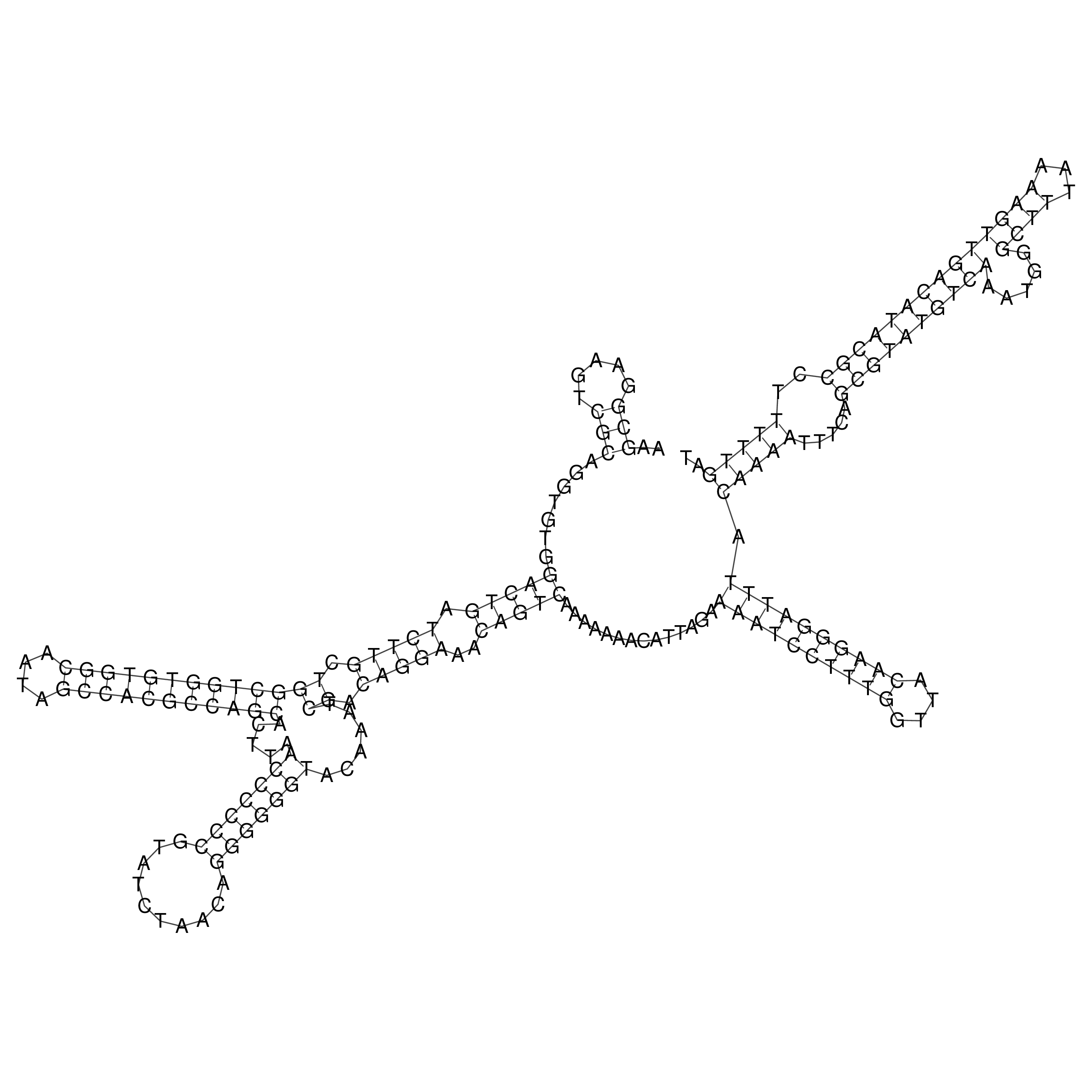
Relaxase
| ID | 2212 | GenBank | WP_000398284 |
| Name | Rep_trans_HTE13_RS00025_P39|unnamed |
UniProt ID | A0A3P3PY43 |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
>WP_000398284.1 MULTISPECIES: MobT family relaxase [Bacteria]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3P3PY43 |
T4CP
| ID | 1895 | GenBank | WP_000813488 |
| Name | tcpA_HTE13_RS00015_P39|unnamed |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>WP_000813488.1 MULTISPECIES: FtsK/SpoIIIE domain-containing protein [Bacteria]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 307..11717
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTE13_RS00005 | 307..621 | + | 315 | WP_000420682 | YdcP family protein | orf23 |
| HTE13_RS00010 | 637..1023 | + | 387 | WP_000985015 | YdcP family protein | orf23 |
| HTE13_RS00015 | 1052..2437 | + | 1386 | WP_000813488 | FtsK/SpoIIIE domain-containing protein | virb4 |
| HTE13_RS00020 | 2440..2592 | + | 153 | WP_000879507 | hypothetical protein | - |
| HTE13_RS00025 | 2615..3820 | + | 1206 | WP_000398284 | MobT family relaxase | - |
| HTE13_RS00030 | 3863..4084 | + | 222 | WP_001009056 | hypothetical protein | orf19 |
| HTE13_RS00035 | 4201..4698 | + | 498 | WP_000342539 | antirestriction protein ArdA | - |
| HTE13_RS00040 | 4787..5179 | + | 393 | WP_000723888 | conjugal transfer protein | orf17a |
| HTE13_RS00045 | 5163..7610 | + | 2448 | WP_000331160 | ATP-binding protein | virb4 |
| HTE13_RS00050 | 7613..9790 | + | 2178 | WP_001574271 | YtxH domain-containing protein | orf15 |
| HTE13_RS00055 | 9787..10788 | + | 1002 | WP_001574272 | bifunctional lysozyme/C40 family peptidase | orf14 |
| HTE13_RS00060 | 10785..11717 | + | 933 | WP_001224319 | conjugal transfer protein | orf13 |
| HTE13_RS00065 | 11962..12078 | + | 117 | WP_001791010 | tetracycline resistance determinant leader peptide | - |
| HTE13_RS00070 | 12094..14013 | + | 1920 | WP_001574275 | tetracycline resistance ribosomal protection protein Tet(M) | - |
| HTE13_RS00080 | 14207..15583 | + | 1377 | WP_001574277 | tetracycline efflux MFS transporter Tet(L) | - |
| HTE13_RS00085 | 15989..16630 | + | 642 | Protein_15 | plasmid recombination protein | - |
Host bacterium
| ID | 3395 | GenBank | NZ_KP345886 |
| Plasmid name | P39|unnamed | Incompatibility group | - |
| Plasmid size | 23910 bp | Coordinate of oriT [Strand] | 2412..2615 [+] |
| Host baterium | Enterococcus faecium isolate P39 |
Cargo genes
| Drug resistance gene | tet(M), tet(L) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |