Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102929 |
| Name | oriT_Tri-48; RCAM 03910|unnamed2 |
| Organism | Phyllobacterium zundukense strain Tri-48 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP017943 (403507..403549 [+], 43 nt) |
| oriT length | 43 nt |
| IRs (inverted repeats) | 21..27, 32..38 (ACGTCGC..GCGACGT) 11..16, 28..33 (GCGCAA..TTGCGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 43 nt
>oriT_Tri-48; RCAM 03910|unnamed2
GGATCCAAGGGCGCAATTATACGTCGCTTGCGCGACGTCCTGC
GGATCCAAGGGCGCAATTATACGTCGCTTGCGCGACGTCCTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file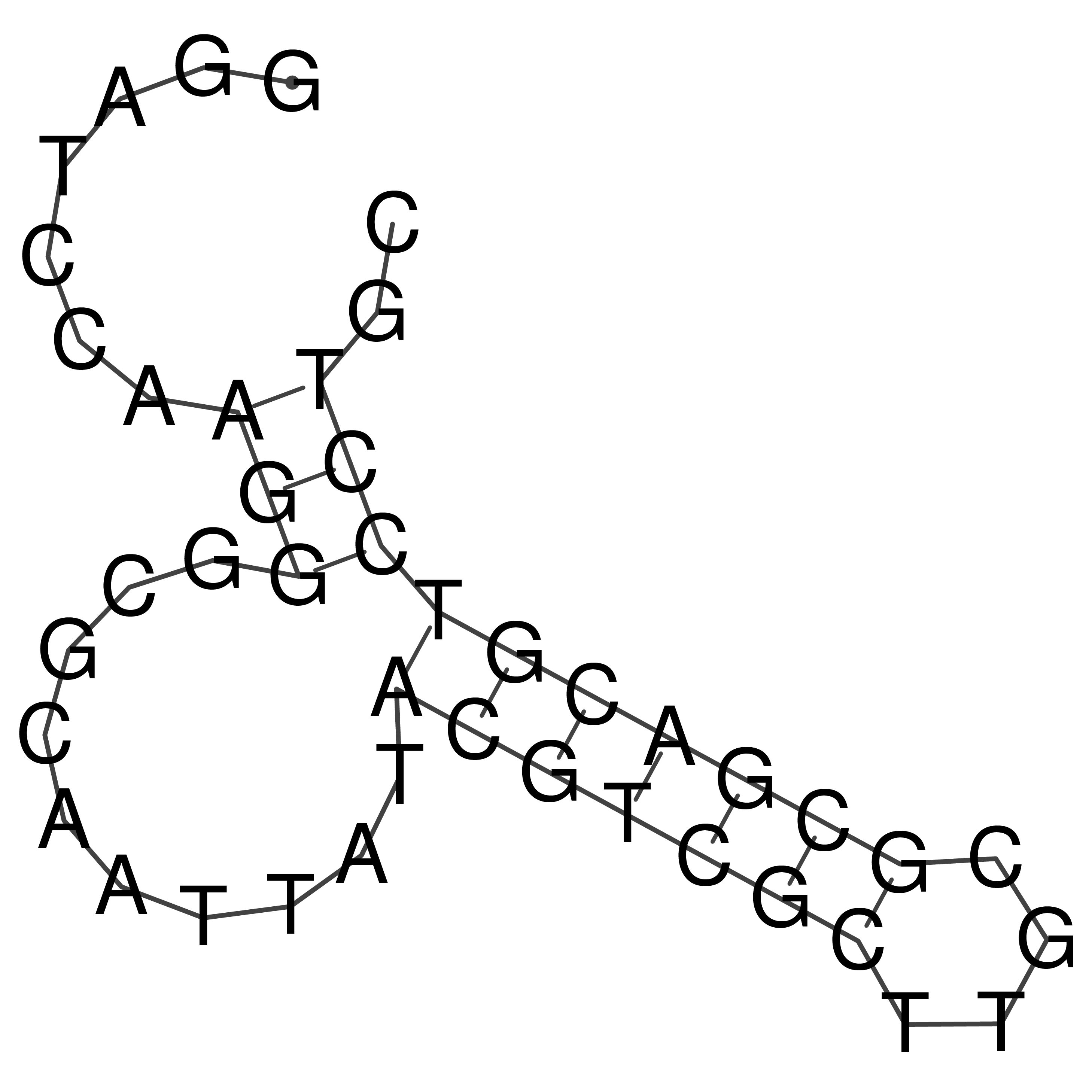
Relaxase
| ID | 2199 | GenBank | WP_100003247 |
| Name | traA_BLM14_RS27630_Tri-48; RCAM 03910|unnamed2 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 122877.38 Da Isoelectric Point: 10.0004
>WP_100003247.1 Ti-type conjugative transfer relaxase TraA [Phyllobacterium zundukense]
MAIAHFSVSIVSRGNGRSAVLSAAYRHCAKMDYEREARTIDYTRKEGLIHEDFSLPADAPDWARTLIADR
STSGASEAFWNKVEAFEKRVDAQLAKDLTVALPLELSVEQNIVLVRDFVERHILSKGMVADWVYHDNPGN
PHIHLMTTLRPLTEDGFGAKKIAVIGEDGQPLRNKTGRQVYQLWAGDTQDFNAFREGWFERQNHHLALNG
IALQVDGRSYEKQGIELVPTIHVGVGAKAIERKAKAEGWKPSLERLDLQEAVRLKNLRRIPARPEIVLDL
VAREKSVFDERDVAKVLHRYVDDPGIFHQLLARILQSPDVLRLQRDTVAFATGEKVPARYTTRELIRLEA
EMAARAIWLSGRSSHGVQKEVLQTTFARHTRLSDEQRAAIEHVMGSARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGAALAGKAAEGLEKEAGIQSRTLASWELRWKQGRDALDARTVFVMDEAGMVASRQMAGFVE
AVVRTGAKLVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQRETWMREASLDLARGNLGQALAAYR
TEGKVLGSELKAEAIETLIADWNRDYDASKTTLILAHLRRDVRLLNEMARTRLVERGIVGEGESFLTADG
MRRFDAGDQIVFLKNDGSLGVKNGMLAKVVEASQNRIVAVVGEGDQRRQVVVEQRFYNNLDHGYATTIHK
SQGATVDCVKVLASLSLDRHLTYVAMTRHREDLQLYYGTRSFAINGGLEKILSRARPKETTLDYERGTLY
RQALRFAENRGLHVVQVARTLLRDRLDWTLRQSSKLADLAVRLRAAGTRLGLLQIPKMDTKKEARPMVSG
VKLFPIPLSDTVEQKLAADPALKTQWEEVSTRFRYVFADPETAFRAMNFDAVLTDGRAASQTLQKLAVNP
VSIGPLNGKTGLLASKSDREARRAANVNVPALKRDIERYLQMREAAVQRLEADEKALRQRVSIDIPALSP
AARSVLERVRDAIDRNDLPAALGYALSNRETKIEIDGFNKAVGERFGERTLLTNAAREPSGKLFDKLADG
LKPEEKQRLRDAWPILRTAQQLAAHERTVVSLKQAEDLRLSQRQTPALKQ
MAIAHFSVSIVSRGNGRSAVLSAAYRHCAKMDYEREARTIDYTRKEGLIHEDFSLPADAPDWARTLIADR
STSGASEAFWNKVEAFEKRVDAQLAKDLTVALPLELSVEQNIVLVRDFVERHILSKGMVADWVYHDNPGN
PHIHLMTTLRPLTEDGFGAKKIAVIGEDGQPLRNKTGRQVYQLWAGDTQDFNAFREGWFERQNHHLALNG
IALQVDGRSYEKQGIELVPTIHVGVGAKAIERKAKAEGWKPSLERLDLQEAVRLKNLRRIPARPEIVLDL
VAREKSVFDERDVAKVLHRYVDDPGIFHQLLARILQSPDVLRLQRDTVAFATGEKVPARYTTRELIRLEA
EMAARAIWLSGRSSHGVQKEVLQTTFARHTRLSDEQRAAIEHVMGSARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGAALAGKAAEGLEKEAGIQSRTLASWELRWKQGRDALDARTVFVMDEAGMVASRQMAGFVE
AVVRTGAKLVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQRETWMREASLDLARGNLGQALAAYR
TEGKVLGSELKAEAIETLIADWNRDYDASKTTLILAHLRRDVRLLNEMARTRLVERGIVGEGESFLTADG
MRRFDAGDQIVFLKNDGSLGVKNGMLAKVVEASQNRIVAVVGEGDQRRQVVVEQRFYNNLDHGYATTIHK
SQGATVDCVKVLASLSLDRHLTYVAMTRHREDLQLYYGTRSFAINGGLEKILSRARPKETTLDYERGTLY
RQALRFAENRGLHVVQVARTLLRDRLDWTLRQSSKLADLAVRLRAAGTRLGLLQIPKMDTKKEARPMVSG
VKLFPIPLSDTVEQKLAADPALKTQWEEVSTRFRYVFADPETAFRAMNFDAVLTDGRAASQTLQKLAVNP
VSIGPLNGKTGLLASKSDREARRAANVNVPALKRDIERYLQMREAAVQRLEADEKALRQRVSIDIPALSP
AARSVLERVRDAIDRNDLPAALGYALSNRETKIEIDGFNKAVGERFGERTLLTNAAREPSGKLFDKLADG
LKPEEKQRLRDAWPILRTAQQLAAHERTVVSLKQAEDLRLSQRQTPALKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1875 | GenBank | WP_100003244 |
| Name | traG_BLM14_RS27615_Tri-48; RCAM 03910|unnamed2 |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 68705.94 Da Isoelectric Point: 9.7841
>WP_100003244.1 Ti-type conjugative transfer system protein TraG [Phyllobacterium zundukense]
MTVNRLVLAIMPATLMITAAIFVPGIEHWLAAFGKTAQAKLMLGRIGLALPYLIVAAIGVIFLFAANGAA
HIKAAGWSVVAGGAATILIAVTREAIRLSGIAGNVPAGQSVLGYADPATMLGASATLFAGVFALRVGLRG
NAAFAKSAPRRIHGKRAVHGEADWMGMTEAAKMFPDAGGIVMGERYRVDRDSVMAVSFRADAQETWGAGG
KAPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLIVLDPSSEVAPMVVDHRRKAGRKVIVLD
PSSPATGFNALDWIGRFGGTKEEDIVAVATWIMTDNARAASARDDFFRASAMQLLTALIADVCLSGHTAE
SEQTLRQVRKNLSEPEPQLRQRLQEIYDNSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGTTFSTSNLAAGNTDVFINIDLKALETHAGLARVIIGAFLNAIYNRDGAMQGRALFLLDEVARLGY
MRILETARDAGRKYGITLIMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRADMKACVG
ANRFQSEKT
MTVNRLVLAIMPATLMITAAIFVPGIEHWLAAFGKTAQAKLMLGRIGLALPYLIVAAIGVIFLFAANGAA
HIKAAGWSVVAGGAATILIAVTREAIRLSGIAGNVPAGQSVLGYADPATMLGASATLFAGVFALRVGLRG
NAAFAKSAPRRIHGKRAVHGEADWMGMTEAAKMFPDAGGIVMGERYRVDRDSVMAVSFRADAQETWGAGG
KAPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLIVLDPSSEVAPMVVDHRRKAGRKVIVLD
PSSPATGFNALDWIGRFGGTKEEDIVAVATWIMTDNARAASARDDFFRASAMQLLTALIADVCLSGHTAE
SEQTLRQVRKNLSEPEPQLRQRLQEIYDNSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGTTFSTSNLAAGNTDVFINIDLKALETHAGLARVIIGAFLNAIYNRDGAMQGRALFLLDEVARLGY
MRILETARDAGRKYGITLIMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRADMKACVG
ANRFQSEKT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1876 | GenBank | WP_100003293 |
| Name | t4cp2_BLM14_RS27905_Tri-48; RCAM 03910|unnamed2 |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 92018.64 Da Isoelectric Point: 5.6597
>WP_100003293.1 conjugal transfer protein TrbE [Phyllobacterium zundukense]
MVALKQFRHSGPSFADLVPYAGLVDNGIMLLKDGSLMAGWYFAGPDSESSTDAERNEISRQINAILARLG
SGWMIQVEAVRVPTTDYPDEADCHFPDAVTRAIDAERRAHFEQERGHFESRHALILTWRPPEPRRSGLTR
YVYSDAGSRSASYADGVFDAFRTSIREIEQYLANIVSIRRMLTQETEERGGFRIARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLVTSELQHSLTPTVENRFLGVVAIDGLPAESWPGILNALDLMPLTYRWSSRFVF
LDDQEARGRLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMTMVAETEDAIAQAASQLVAYGYYTPVIVL
FDDEQPRLQEKCEAVRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLVPLNSVWS
GSPLAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYRDAQLFA
FDKGRSMLPLTLAARGDHYEIGGDAGGEGDGGSPRLAFCPLAELSSDGDRAWAAEWLETLVALQGVMVTP
DHRNAISRQIALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLRLGAFQCFEVEEL
MNMGERNLVPVLTYLFRRVEKRLTGAPSLIILDEAWLMLGHPLFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREHYVASPDGRRLFDM
ALGPVALSFVGASGKEDLKRIRALHFEHGDNWPEHWLKQRGIAHAQIDLPIG
MVALKQFRHSGPSFADLVPYAGLVDNGIMLLKDGSLMAGWYFAGPDSESSTDAERNEISRQINAILARLG
SGWMIQVEAVRVPTTDYPDEADCHFPDAVTRAIDAERRAHFEQERGHFESRHALILTWRPPEPRRSGLTR
YVYSDAGSRSASYADGVFDAFRTSIREIEQYLANIVSIRRMLTQETEERGGFRIARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLVTSELQHSLTPTVENRFLGVVAIDGLPAESWPGILNALDLMPLTYRWSSRFVF
LDDQEARGRLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMTMVAETEDAIAQAASQLVAYGYYTPVIVL
FDDEQPRLQEKCEAVRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLVPLNSVWS
GSPLAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYRDAQLFA
FDKGRSMLPLTLAARGDHYEIGGDAGGEGDGGSPRLAFCPLAELSSDGDRAWAAEWLETLVALQGVMVTP
DHRNAISRQIALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLRLGAFQCFEVEEL
MNMGERNLVPVLTYLFRRVEKRLTGAPSLIILDEAWLMLGHPLFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREHYVASPDGRRLFDM
ALGPVALSFVGASGKEDLKRIRALHFEHGDNWPEHWLKQRGIAHAQIDLPIG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 460827..470446
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BLM14_RS27850 (BLM14_27270) | 456869..457507 | - | 639 | WP_100003285 | dethiobiotin synthase | - |
| BLM14_RS27855 (BLM14_27275) | 457504..458649 | - | 1146 | WP_100003286 | 8-amino-7-oxononanoate synthase | - |
| BLM14_RS27860 (BLM14_27280) | 458646..459640 | - | 995 | Protein_438 | biotin synthase BioB | - |
| BLM14_RS27865 (BLM14_27285) | 459806..460473 | - | 668 | Protein_439 | GntR family transcriptional regulator | - |
| BLM14_RS27870 (BLM14_27290) | 460827..462137 | - | 1311 | WP_100003287 | IncP-type conjugal transfer protein TrbI | virB10 |
| BLM14_RS27875 (BLM14_27295) | 462150..462611 | - | 462 | WP_100003288 | conjugal transfer protein TrbH | - |
| BLM14_RS27880 (BLM14_27300) | 462611..463456 | - | 846 | WP_100003425 | P-type conjugative transfer protein TrbG | virB9 |
| BLM14_RS27885 (BLM14_27305) | 463474..464136 | - | 663 | WP_100003289 | conjugal transfer protein TrbF | virB8 |
| BLM14_RS27890 (BLM14_27310) | 464155..465336 | - | 1182 | WP_100003290 | P-type conjugative transfer protein TrbL | virB6 |
| BLM14_RS27895 (BLM14_27315) | 465330..465530 | - | 201 | WP_100003291 | entry exclusion protein TrbK | - |
| BLM14_RS27900 (BLM14_27320) | 465527..466267 | - | 741 | WP_237143722 | P-type conjugative transfer protein TrbJ | virB5 |
| BLM14_RS27905 (BLM14_27325) | 466308..468776 | - | 2469 | WP_100003293 | conjugal transfer protein TrbE | virb4 |
| BLM14_RS27910 (BLM14_27330) | 468786..469085 | - | 300 | WP_100003294 | conjugal transfer protein TrbD | virB3 |
| BLM14_RS27915 (BLM14_27335) | 469078..469485 | - | 408 | WP_100003295 | TrbC/VirB2 family protein | virB2 |
| BLM14_RS27920 (BLM14_27340) | 469475..470446 | - | 972 | WP_100003296 | P-type conjugative transfer ATPase TrbB | virB11 |
| BLM14_RS27925 (BLM14_27345) | 470443..471081 | - | 639 | WP_100003297 | acyl-homoserine-lactone synthase TraI | - |
| BLM14_RS27930 (BLM14_27350) | 471421..472644 | + | 1224 | WP_162293266 | plasmid partitioning protein RepA | - |
| BLM14_RS27935 (BLM14_27355) | 472641..473672 | + | 1032 | WP_100003299 | plasmid partitioning protein RepB | - |
| BLM14_RS27940 (BLM14_27360) | 473866..475080 | + | 1215 | WP_100003300 | plasmid replication protein RepC | - |
Host bacterium
| ID | 3372 | GenBank | NZ_CP017943 |
| Plasmid name | Tri-48; RCAM 03910|unnamed2 | Incompatibility group | - |
| Plasmid size | 586378 bp | Coordinate of oriT [Strand] | 403507..403549 [+] |
| Host baterium | Phyllobacterium zundukense strain Tri-48 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixA, fixB |
| Anti-CRISPR | AcrIF11 |