Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102926 |
| Name | oriT_ZLB004|p3 |
| Organism | Levilactobacillus brevis strain ZLB004 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP021459 (40538..40741 [+], 204 nt) |
| oriT length | 204 nt |
| IRs (inverted repeats) | 125..130, 143..148 (AAATCC..GGATTT) 66..72, 83..89 (ACCCCCC..GGGGGGT) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 204 nt
>oriT_ZLB004|p3
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file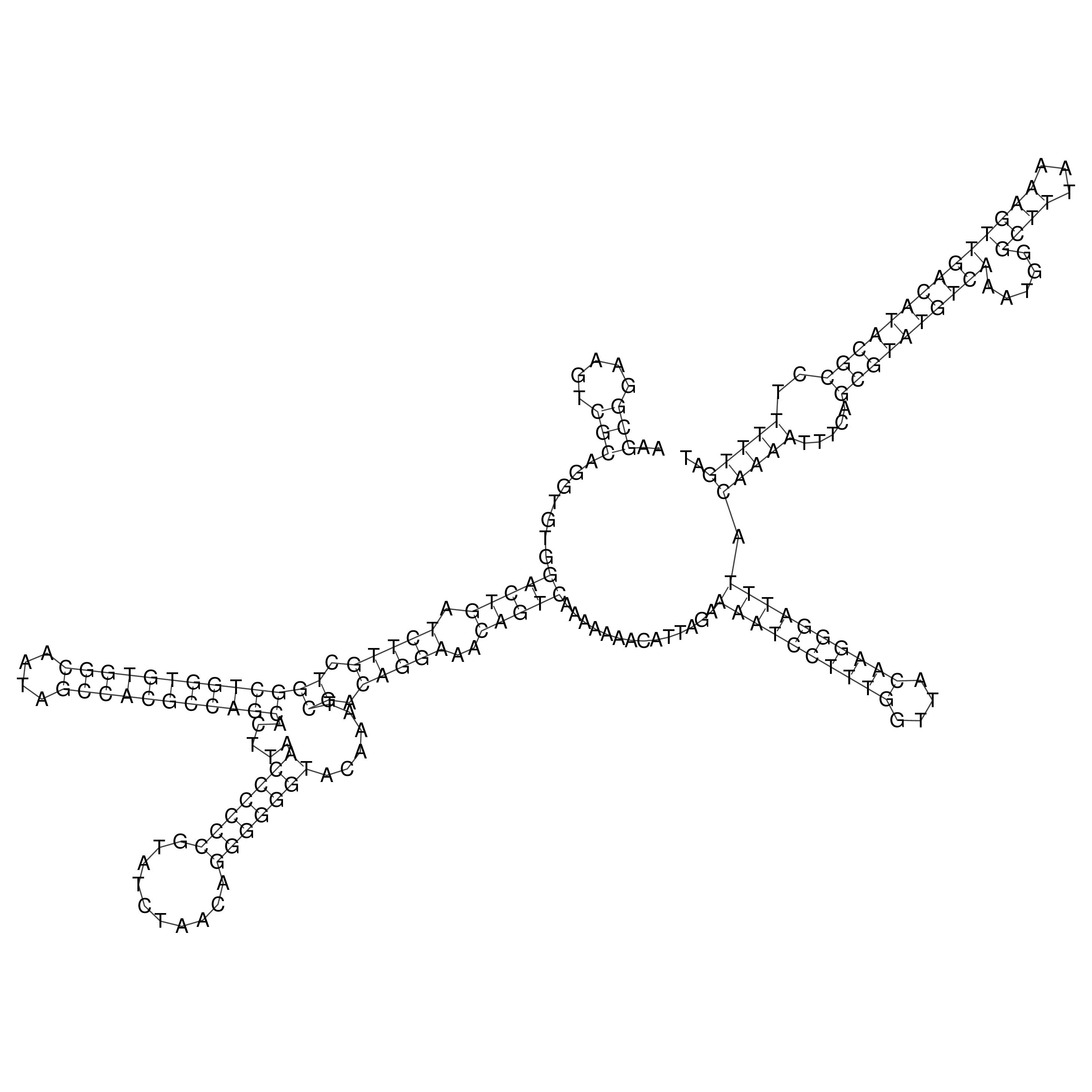
Relaxase
| ID | 2196 | GenBank | WP_000398284 |
| Name | Rep_trans_CCS05_RS12675_ZLB004|p3 |
UniProt ID | A0A3P3PY43 |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
>WP_000398284.1 MULTISPECIES: MobT family relaxase [Bacteria]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3P3PY43 |
T4CP
| ID | 1873 | GenBank | WP_000813488 |
| Name | tcpA_CCS05_RS12665_ZLB004|p3 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>WP_000813488.1 MULTISPECIES: FtsK/SpoIIIE domain-containing protein [Bacteria]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 30993..49843
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CCS05_RS12590 (CCS05_12595) | 26498..27496 | - | 999 | WP_012490951 | IS30 family transposase | - |
| CCS05_RS12595 (CCS05_12600) | 27720..28889 | + | 1170 | WP_110140126 | metallophosphoesterase | - |
| CCS05_RS12600 (CCS05_12605) | 28901..29257 | + | 357 | Protein_33 | asparagine synthetase B | - |
| CCS05_RS12610 (CCS05_12615) | 29432..30670 | - | 1239 | WP_110140127 | ISL3 family transposase | - |
| CCS05_RS12615 (CCS05_12620) | 30993..31922 | + | 930 | WP_110140128 | ABC transporter ATP-binding protein | virb4 |
| CCS05_RS12620 (CCS05_12625) | 31924..33444 | + | 1521 | WP_110140129 | ABC transporter permease/substrate-binding protein | - |
| CCS05_RS12625 (CCS05_12630) | 33847..34776 | - | 930 | WP_110140130 | IS30-like element ISLpl1 family transposase | - |
| CCS05_RS12635 (CCS05_12635) | 35437..36018 | + | 582 | WP_014216283 | recombinase family protein | - |
| CCS05_RS12640 (CCS05_12640) | 36416..37048 | + | 633 | WP_110140131 | recombinase family protein | - |
| CCS05_RS12645 (CCS05_12645) | 37151..38002 | + | 852 | WP_225426374 | Fic family protein | - |
| CCS05_RS12655 (CCS05_12655) | 38433..38747 | + | 315 | WP_000420682 | YdcP family protein | orf23 |
| CCS05_RS12660 (CCS05_12660) | 38763..39149 | + | 387 | WP_000985015 | YdcP family protein | orf23 |
| CCS05_RS12665 (CCS05_12665) | 39178..40563 | + | 1386 | WP_000813488 | FtsK/SpoIIIE domain-containing protein | virb4 |
| CCS05_RS12670 (CCS05_12670) | 40566..40718 | + | 153 | WP_000879507 | hypothetical protein | - |
| CCS05_RS12675 (CCS05_12675) | 40741..41946 | + | 1206 | WP_000398284 | MobT family relaxase | - |
| CCS05_RS12680 (CCS05_12680) | 41989..42210 | + | 222 | WP_001009056 | hypothetical protein | orf19 |
| CCS05_RS12685 (CCS05_12685) | 42327..42824 | + | 498 | WP_000342539 | antirestriction protein ArdA | - |
| CCS05_RS12690 (CCS05_12690) | 42913..43305 | + | 393 | WP_240005880 | conjugal transfer protein | orf17a |
| CCS05_RS12695 (CCS05_12695) | 43289..45736 | + | 2448 | WP_000331160 | ATP-binding protein | virb4 |
| CCS05_RS12700 (CCS05_12700) | 45739..47916 | + | 2178 | WP_000804748 | membrane protein | orf15 |
| CCS05_RS12705 (CCS05_12705) | 47913..48914 | + | 1002 | WP_000769868 | bifunctional lysozyme/C40 family peptidase | orf14 |
| CCS05_RS12710 (CCS05_12710) | 48911..49843 | + | 933 | WP_001224319 | conjugal transfer protein | orf13 |
| CCS05_RS13510 (CCS05_12715) | 50088..50204 | + | 117 | WP_001791010 | tetracycline resistance determinant leader peptide | - |
| CCS05_RS12720 (CCS05_12720) | 50220..52139 | + | 1920 | WP_110140133 | tetracycline resistance ribosomal protection protein Tet(M) | - |
Host bacterium
| ID | 3369 | GenBank | NZ_CP021459 |
| Plasmid name | ZLB004|p3 | Incompatibility group | - |
| Plasmid size | 52778 bp | Coordinate of oriT [Strand] | 40538..40741 [+] |
| Host baterium | Levilactobacillus brevis strain ZLB004 |
Cargo genes
| Drug resistance gene | tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |