Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102923 |
| Name | oriT_p1540-4 |
| Organism | Escherichia coli strain CRE1540 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP019055 (153353..153704 [+], 352 nt) |
| oriT length | 352 nt |
| IRs (inverted repeats) | 255..262, 271..278 (ACCGCTAG..CTAGCGGT) 192..198, 206..212 (TATAAAA..TTTTATA) 40..47, 50..57 (GCAAAAAC..GTTTTTGC) 4..11, 16..23 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 352 nt
AGGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTTTCTTTATAAATAGAGAGTTATGAAAAATTAGTTTCTCTTACTCTCTTTATGATATTTAAAAAAGCGGTGTCGGCGAGGCTACAACAACGCGCCGACACCGCTTTGTAGGGGTGGTACTGACTATTTTTATAAAAAACATTATTTTATATTAGGGGTGCTGCTAGCGGCGCGGTGTGTTTTTTTATAGGATACCGCTAGGGGCGCTGCTAGCGGTGCGTCCCTGTTTGCATTATGAATTTTAGTGTTTCGAAATTAACTTTATTTTATGTTCAAAAAAGGCAATCTCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file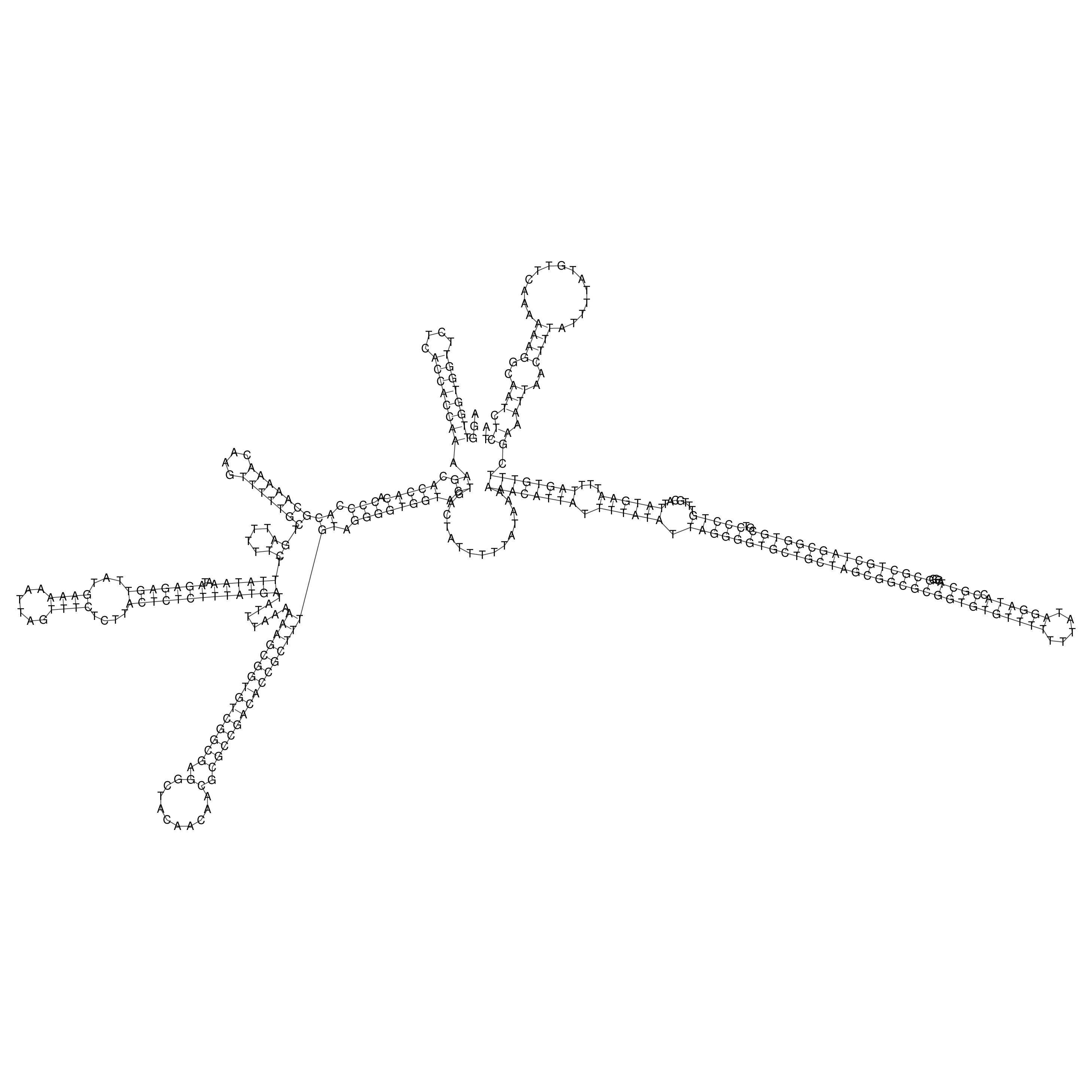
Relaxase
| ID | 2194 | GenBank | WP_001611029 |
| Name | traI_BVL39_RS28675_p1540-4 |
UniProt ID | A0A075MCD1 |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191983.83 Da Isoelectric Point: 5.9594
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGKLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQTPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQEGMAFTPGNTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGLPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFTALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTSFIDLGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWVPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDIPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRECGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMTGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKSDREVMNAERLFSTARELRDVVAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPRTITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVRDIAGLERDRSAISEREAALPESVLREPQRVREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 897 | GenBank | WP_001151524 |
| Name | WP_001151524_p1540-4 |
UniProt ID | A0A5W2DT22 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14517.48 Da Isoelectric Point: 5.0516
MAKVNLYISNDAYEKINAIIEKRRQEGAREKDVSFSATASMLLELGLRVHEAQMERKESAFNQTEFNKLL
LECVVKTQSSVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSSEMERFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A5W2DT22 |
| ID | 898 | GenBank | WP_001369361 |
| Name | WP_001369361_p1540-4 |
UniProt ID | B7LI64 |
| Length | 133 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 133 a.a. Molecular weight: 15409.70 Da Isoelectric Point: 9.9240
MLLKRFGTRSATGKMVKLKLPVDVESLLIEASNRSGRSRSFEAVIRLKDHLHRYPKFNRAGNIYGKSLVK
YLTMRLDDETNQLLIAAKNRSGWCKTDEAADRVIDHLIKFPDFYNSEIFREADKEEDITFNTL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B7LI64 |
T4CP
| ID | 1870 | GenBank | WP_021526192 |
| Name | traD_BVL39_RS28660_p1540-4 |
UniProt ID | _ |
| Length | 735 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 735 a.a. Molecular weight: 83648.41 Da Isoelectric Point: 5.1126
MSFNAKDMTQGGQIASMRIRMFSQIANIILYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIRSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLASVVALVICLITFFVVSWILGRQGKQ
QSENEITGGRQLTDNPKEVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQTRPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPQQPVSPAINDKKSDAGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAVYEAWQ
REQNPDIQQKMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 152782..165375
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BVL39_RS28555 (BVL39_29035) | 147932..149473 | - | 1542 | WP_001298859 | IS21-like element ISEc12 family transposase | - |
| BVL39_RS28560 (BVL39_29040) | 149666..150236 | + | 571 | Protein_158 | plasmid SOS inhibition protein A | - |
| BVL39_RS29815 | 150415..150564 | + | 150 | Protein_159 | plasmid maintenance protein Mok | - |
| BVL39_RS28565 (BVL39_29050) | 150506..150631 | + | 126 | WP_096937776 | type I toxin-antitoxin system Hok family toxin | - |
| BVL39_RS29820 | 150932..151205 | - | 274 | Protein_161 | hypothetical protein | - |
| BVL39_RS28575 (BVL39_29060) | 151257..151553 | + | 297 | WP_001272245 | hypothetical protein | - |
| BVL39_RS28580 (BVL39_29065) | 151664..152485 | + | 822 | WP_001234487 | DUF932 domain-containing protein | - |
| BVL39_RS28585 (BVL39_29070) | 152782..153372 | - | 591 | WP_279947734 | transglycosylase SLT domain-containing protein | - |
| BVL39_RS28590 (BVL39_29075) | 153705..154088 | + | 384 | WP_001151524 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| BVL39_RS28595 (BVL39_29080) | 154275..154964 | + | 690 | WP_000283385 | conjugal transfer transcriptional regulator TraJ | - |
| BVL39_RS28600 (BVL39_29085) | 155063..155458 | + | 396 | WP_001309237 | conjugal transfer relaxosome DNA-bindin protein TraY | - |
| BVL39_RS28605 (BVL39_29090) | 155491..155856 | + | 366 | WP_000994779 | type IV conjugative transfer system pilin TraA | - |
| BVL39_RS28610 (BVL39_29095) | 155871..156182 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| BVL39_RS28615 (BVL39_29100) | 156204..156770 | + | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| BVL39_RS28620 (BVL39_29105) | 156757..157485 | + | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| BVL39_RS28625 (BVL39_29110) | 157485..158912 | + | 1428 | WP_000146685 | F-type conjugal transfer pilus assembly protein TraB | traB |
| BVL39_RS28630 (BVL39_29115) | 158902..159492 | + | 591 | WP_000002787 | conjugal transfer pilus-stabilizing protein TraP | - |
| BVL39_RS28635 (BVL39_29120) | 159479..159676 | + | 198 | WP_001324648 | conjugal transfer protein TrbD | - |
| BVL39_RS28640 (BVL39_29125) | 159688..159939 | + | 252 | WP_001038342 | conjugal transfer protein TrbG | - |
| BVL39_RS29375 | 159936..160370 | + | 435 | Protein_176 | type IV conjugative transfer system lipoprotein TraV | - |
| BVL39_RS28645 (BVL39_29130) | 160371..161639 | + | 1269 | Protein_177 | conjugal transfer protein TraG | - |
| BVL39_RS28650 (BVL39_29135) | 161655..162152 | + | 498 | WP_000605857 | entry exclusion protein | - |
| BVL39_RS28655 (BVL39_29140) | 162184..162915 | + | 732 | WP_023565183 | conjugal transfer complement resistance protein TraT | - |
| BVL39_RS28660 (BVL39_29145) | 163168..165375 | + | 2208 | WP_021526192 | type IV conjugative transfer system coupling protein TraD | virb4 |
| BVL39_RS28665 (BVL39_29150) | 165384..165782 | - | 399 | WP_000911313 | type II toxin-antitoxin system VapC family toxin | - |
| BVL39_RS28670 (BVL39_29155) | 165782..166009 | - | 228 | WP_000450520 | toxin-antitoxin system antitoxin VapB | - |
Host bacterium
| ID | 3366 | GenBank | NZ_CP019055 |
| Plasmid name | p1540-4 | Incompatibility group | IncFIB |
| Plasmid size | 184166 bp | Coordinate of oriT [Strand] | 153353..153704 [+] |
| Host baterium | Escherichia coli strain CRE1540 |
Cargo genes
| Drug resistance gene | sitABCD |
| Virulence gene | vat, iroN, iroE, iroD, iroC, iroB, iucA, iucB, iucC, iutA |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |