Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102899 |
| Name | oriT_pSE79-2359 |
| Organism | Salmonella enterica subsp. enterica serovar Enteritidis strain 79-2359 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP018646 (49814..49914 [-], 101 nt) |
| oriT length | 101 nt |
| IRs (inverted repeats) | 80..85, 91..96 (AAAAAA..TTTTTT) 20..26, 38..44 (TAAATCA..TGATTTA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 101 nt
>oriT_pSE79-2359
TATTTATTTTTTTATTTTTTAAATCAGTGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
TATTTATTTTTTTATTTTTTAAATCAGTGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file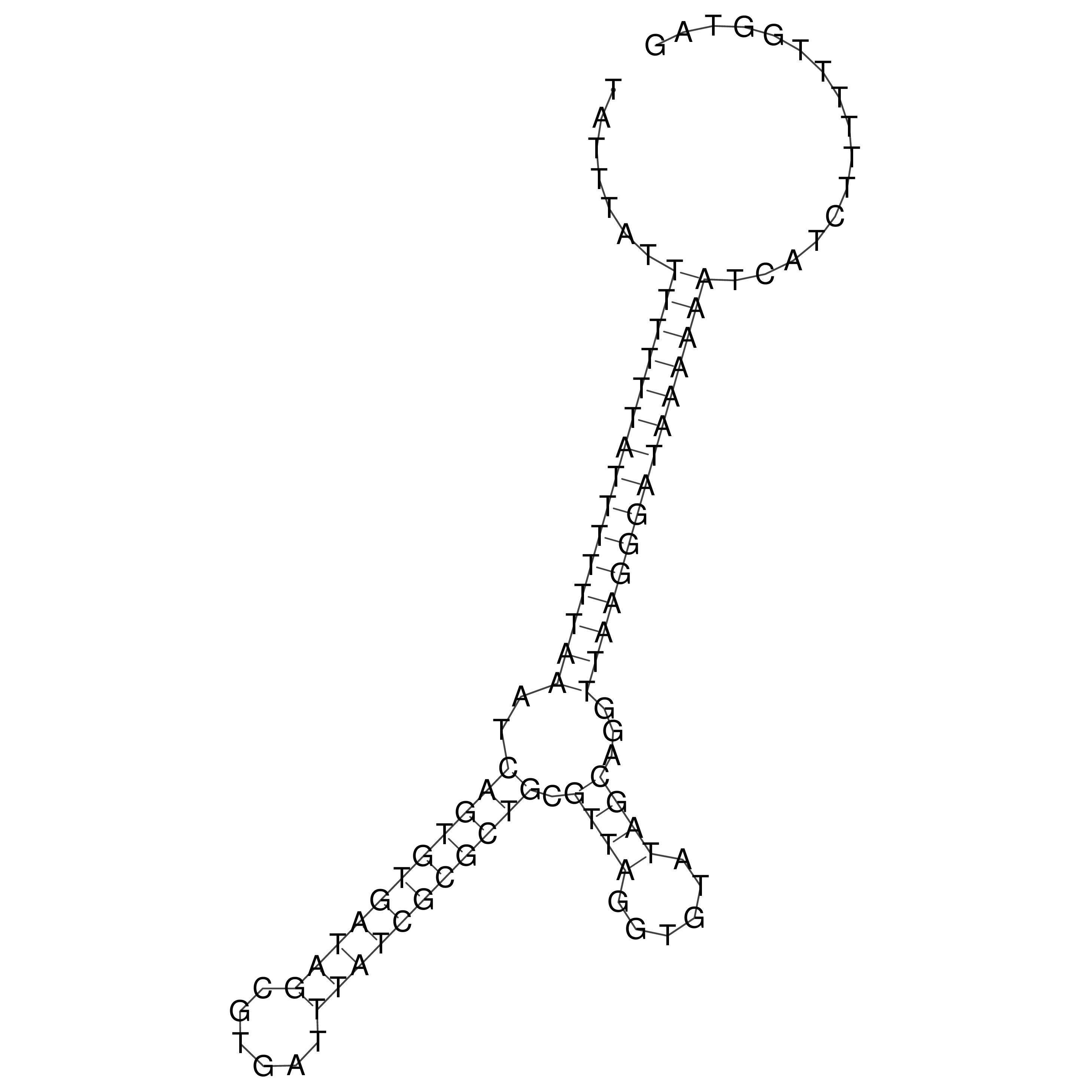
Relaxase
| ID | 2177 | GenBank | WP_000884352 |
| Name | TrwC_BTN70_RS23895_pSE79-2359 |
UniProt ID | A8R732 |
| Length | 1078 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1078 a.a. Molecular weight: 120163.41 Da Isoelectric Point: 6.5766
>WP_000884352.1 MULTISPECIES: MobF family relaxase [Enterobacterales]
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLAGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEEGGHEI
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLAGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEEGGHEI
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A8R732 |
Auxiliary protein
| ID | 886 | GenBank | WP_000706094 |
| Name | WP_000706094_pSE79-2359 |
UniProt ID | B6UZ32 |
| Length | 96 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 96 a.a. Molecular weight: 10829.54 Da Isoelectric Point: 8.5040
>WP_000706094.1 MULTISPECIES: hypothetical protein [Enterobacterales]
MKIVADRLSDVNRKLDYLFDRASDADFGPLRDELKAITETLSGVKFPPAGQMMLHESLAIETLILLRSIA
EPGKTKAAKAEVERNGYKVWEPKKER
MKIVADRLSDVNRKLDYLFDRASDADFGPLRDELKAITETLSGVKFPPAGQMMLHESLAIETLILLRSIA
EPGKTKAAKAEVERNGYKVWEPKKER
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B6UZ32 |
T4CP
| ID | 1849 | GenBank | WP_000342688 |
| Name | t4cp2_BTN70_RS24185_pSE79-2359 |
UniProt ID | _ |
| Length | 509 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 509 a.a. Molecular weight: 57762.87 Da Isoelectric Point: 9.6551
>WP_000342688.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Enterobacterales]
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVRARTLADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVKYHRKNPVPGIELREI
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVRARTLADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVKYHRKNPVPGIELREI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 12383..22708
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BTN70_RS23925 (BTN70_23870) | 7640..8200 | + | 561 | WP_001161490 | recombinase family protein | - |
| BTN70_RS23930 (BTN70_23875) | 8204..11170 | + | 2967 | WP_001138014 | Tn3-like element TnAs1 family transposase | - |
| BTN70_RS24830 | 11281..11676 | + | 396 | WP_306766596 | DUF6710 family protein | - |
| BTN70_RS23940 (BTN70_23885) | 11850..12383 | - | 534 | WP_000731968 | phospholipase D family protein | - |
| BTN70_RS23945 (BTN70_23890) | 12383..13378 | - | 996 | WP_000128596 | ATPase, T2SS/T4P/T4SS family | virB11 |
| BTN70_RS23950 (BTN70_23895) | 13420..14580 | - | 1161 | WP_000101711 | type IV secretion system protein VirB10 | virB10 |
| BTN70_RS23955 (BTN70_23900) | 14580..15464 | - | 885 | WP_000735067 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| BTN70_RS23960 (BTN70_23905) | 15475..16173 | - | 699 | WP_000646594 | type IV secretion system protein | virB8 |
| BTN70_RS24835 (BTN70_23910) | 16163..16321 | - | 159 | WP_012561180 | hypothetical protein | - |
| BTN70_RS23970 (BTN70_23915) | 16392..17432 | - | 1041 | WP_000886022 | type IV secretion system protein | virB6 |
| BTN70_RS23975 (BTN70_23920) | 17448..17675 | - | 228 | WP_000734973 | IncN-type entry exclusion lipoprotein EexN | - |
| BTN70_RS23980 (BTN70_23925) | 17683..18396 | - | 714 | WP_000749362 | type IV secretion system protein | virB5 |
| BTN70_RS23985 (BTN70_23930) | 18414..21014 | - | 2601 | WP_001200711 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| BTN70_RS23990 (BTN70_23935) | 21014..21331 | - | 318 | WP_000496058 | VirB3 family type IV secretion system protein | virB3 |
| BTN70_RS23995 (BTN70_23940) | 21381..21674 | - | 294 | WP_000209137 | TrbC/VirB2 family protein | virB2 |
| BTN70_RS24000 (BTN70_23945) | 21684..21965 | - | 282 | WP_000440698 | transcriptional repressor KorA | - |
| BTN70_RS24005 (BTN70_23950) | 21974..22708 | - | 735 | WP_000033783 | lytic transglycosylase domain-containing protein | virB1 |
| BTN70_RS24010 (BTN70_23955) | 22817..23122 | + | 306 | WP_000960954 | H-NS family nucleoid-associated regulatory protein | - |
| BTN70_RS24710 | 23138..23314 | + | 177 | WP_232516986 | hypothetical protein | - |
| BTN70_RS24020 (BTN70_23965) | 23477..23791 | + | 315 | WP_000734776 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| BTN70_RS24025 (BTN70_23970) | 23827..24138 | + | 312 | WP_001452736 | hypothetical protein | - |
| BTN70_RS24030 (BTN70_23980) | 24264..25100 | - | 837 | WP_000480968 | aminoglycoside O-phosphotransferase APH(6)-Id | - |
| BTN70_RS24035 (BTN70_23985) | 25100..25903 | - | 804 | WP_001082319 | aminoglycoside O-phosphotransferase APH(3'')-Ib | - |
Host bacterium
| ID | 3342 | GenBank | NZ_CP018646 |
| Plasmid name | pSE79-2359 | Incompatibility group | IncN |
| Plasmid size | 54796 bp | Coordinate of oriT [Strand] | 49814..49914 [-] |
| Host baterium | Salmonella enterica subsp. enterica serovar Enteritidis strain 79-2359 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |