Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102897 |
| Name | oriT_pSE74-1357 |
| Organism | Salmonella enterica subsp. enterica serovar Enteritidis strain 74-1357 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP018643 (111288..111388 [+], 101 nt) |
| oriT length | 101 nt |
| IRs (inverted repeats) | 80..85, 91..96 (AAAAAA..TTTTTT) 20..26, 38..44 (TAAATCA..TGATTTA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 101 nt
TATTTATTTTTTTATTTTTTAAATCAGTGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file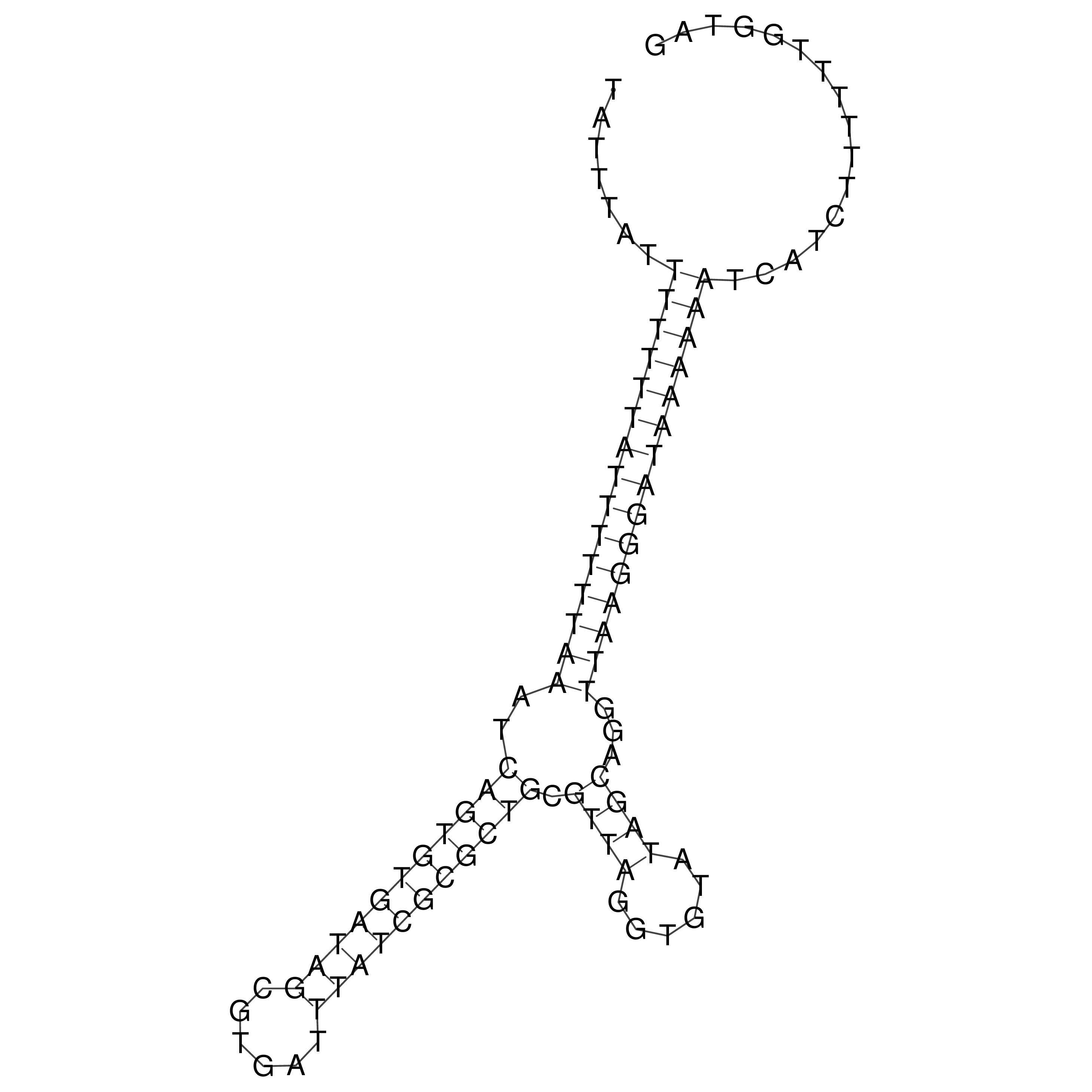
Relaxase
| ID | 2175 | GenBank | WP_097478309 |
| Name | TrwC_BTN71_RS24705_pSE74-1357 |
UniProt ID | _ |
| Length | 1078 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1078 a.a. Molecular weight: 120193.44 Da Isoelectric Point: 6.5766
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGETRYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLAGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEEGGHEI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 881 | GenBank | WP_000089263 |
| Name | WP_000089263_pSE74-1357 |
UniProt ID | A0A3U3XJL9 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 8541.74 Da Isoelectric Point: 8.6796
MSRNIIRPAPGNKVLLVLDDATNHKLLGARERSGRTKTNEVLVRLRDHLNRFPDFYNLDAIKEGAEETDS
IIKDL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3U3XJL9 |
| ID | 882 | GenBank | WP_001354030 |
| Name | WP_001354030_pSE74-1357 |
UniProt ID | A0A734M5H3 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14508.47 Da Isoelectric Point: 4.7116
MARVILYISNDVYDKVNAIVEQRRQEGARDKDISVSGTASMLLELGLRVYEAQMERKESAFNQTEFNKLL
LECVVKTQSSVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSSEMERFFPKNDEE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A734M5H3 |
| ID | 883 | GenBank | WP_000706094 |
| Name | WP_000706094_pSE74-1357 |
UniProt ID | B6UZ32 |
| Length | 96 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 96 a.a. Molecular weight: 10829.54 Da Isoelectric Point: 8.5040
MKIVADRLSDVNRKLDYLFDRASDADFGPLRDELKAITETLSGVKFPPAGQMMLHESLAIETLILLRSIA
EPGKTKAAKAEVERNGYKVWEPKKER
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B6UZ32 |
T4CP
| ID | 1846 | GenBank | WP_000069777 |
| Name | traC_BTN71_RS24255_pSE74-1357 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 99195.86 Da Isoelectric Point: 6.1126
MSNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAI
PINGANKSIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAA
ATQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASIATQTVDAQAFIDIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLNVRADYLTLGLRENGRNSTARILNFHLARNPEIAF
LWNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAGSPTIRSRLDEMIVLLDQY
TANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGW
RLLDFKNRKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKY
NQLYPDQFQPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEG
LSIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1847 | GenBank | WP_000342688 |
| Name | t4cp2_BTN71_RS24710_pSE74-1357 |
UniProt ID | _ |
| Length | 509 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 509 a.a. Molecular weight: 57762.87 Da Isoelectric Point: 9.6551
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVRARTLADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVKYHRKNPVPGIELREI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 26370..50383
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BTN71_RS24145 (BTN71_23990) | 21910..22122 | - | 213 | WP_001309245 | ANR family transcriptional regulator | - |
| BTN71_RS24150 (BTN71_23995) | 22251..22811 | - | 561 | WP_000139323 | fertility inhibition protein FinO | - |
| BTN71_RS24155 (BTN71_24000) | 22914..23774 | - | 861 | WP_000704523 | alpha/beta hydrolase | - |
| BTN71_RS24160 (BTN71_24005) | 23833..24078 | - | 246 | Protein_29 | TraX family protein | - |
| BTN71_RS24165 (BTN71_24010) | 24149..24886 | - | 738 | WP_000199905 | hypothetical protein | - |
| BTN71_RS24170 (BTN71_24015) | 25089..25820 | - | 732 | WP_000782451 | conjugal transfer complement resistance protein TraT | - |
| BTN71_RS24175 (BTN71_24020) | 25869..26354 | - | 486 | WP_000605870 | hypothetical protein | - |
| BTN71_RS24180 (BTN71_24025) | 26370..29192 | - | 2823 | WP_001007039 | conjugal transfer mating-pair stabilization protein TraG | traG |
| BTN71_RS24185 (BTN71_24030) | 29189..30562 | - | 1374 | WP_000944331 | conjugal transfer pilus assembly protein TraH | traH |
| BTN71_RS24190 (BTN71_24035) | 30549..30941 | - | 393 | WP_000660699 | F-type conjugal transfer protein TrbF | - |
| BTN71_RS24195 (BTN71_24040) | 30922..31269 | - | 348 | WP_001309242 | P-type conjugative transfer protein TrbJ | - |
| BTN71_RS24200 (BTN71_24045) | 31199..31744 | - | 546 | WP_000059831 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| BTN71_RS24205 (BTN71_24050) | 31731..32015 | - | 285 | WP_000624194 | type-F conjugative transfer system pilin chaperone TraQ | - |
| BTN71_RS24210 (BTN71_24055) | 32134..32478 | - | 345 | WP_000556796 | conjugal transfer protein TrbA | - |
| BTN71_RS24215 (BTN71_24060) | 32492..33235 | - | 744 | WP_001030371 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| BTN71_RS24220 (BTN71_24065) | 33228..33485 | - | 258 | WP_000864353 | conjugal transfer protein TrbE | - |
| BTN71_RS24225 (BTN71_24070) | 33512..35362 | - | 1851 | WP_000821856 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| BTN71_RS24230 (BTN71_24075) | 35359..35997 | - | 639 | WP_001080256 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| BTN71_RS24235 (BTN71_24080) | 36006..36311 | - | 306 | WP_000224416 | hypothetical protein | - |
| BTN71_RS24240 (BTN71_24085) | 36341..37333 | - | 993 | WP_000830838 | conjugal transfer pilus assembly protein TraU | traU |
| BTN71_RS24245 (BTN71_24090) | 37330..37962 | - | 633 | WP_001203728 | type-F conjugative transfer system protein TraW | traW |
| BTN71_RS24250 (BTN71_24095) | 37959..38345 | - | 387 | WP_000214084 | type-F conjugative transfer system protein TrbI | - |
| BTN71_RS24255 (BTN71_24100) | 38342..40972 | - | 2631 | WP_000069777 | type IV secretion system protein TraC | virb4 |
| BTN71_RS24260 (BTN71_24105) | 41098..41445 | - | 348 | WP_000836682 | hypothetical protein | - |
| BTN71_RS24265 (BTN71_24110) | 41473..41691 | - | 219 | WP_000556745 | hypothetical protein | - |
| BTN71_RS24270 (BTN71_24115) | 41771..42244 | - | 474 | WP_000549568 | hypothetical protein | - |
| BTN71_RS24275 (BTN71_24120) | 42237..42650 | - | 414 | WP_000549589 | hypothetical protein | - |
| BTN71_RS24280 (BTN71_24125) | 42643..42864 | - | 222 | WP_001278683 | conjugal transfer protein TraR | - |
| BTN71_RS24285 (BTN71_24130) | 42999..43514 | - | 516 | WP_000809881 | type IV conjugative transfer system lipoprotein TraV | traV |
| BTN71_RS24290 (BTN71_24135) | 43511..43831 | - | 321 | WP_001057307 | conjugal transfer protein TrbD | - |
| BTN71_RS24295 (BTN71_24140) | 43818..44384 | - | 567 | WP_000896599 | conjugal transfer pilus-stabilizing protein TraP | - |
| BTN71_RS24300 (BTN71_24145) | 44374..45825 | - | 1452 | WP_000146675 | F-type conjugal transfer pilus assembly protein TraB | traB |
| BTN71_RS24305 (BTN71_24150) | 45825..46553 | - | 729 | WP_001230772 | type-F conjugative transfer system secretin TraK | traK |
| BTN71_RS24310 (BTN71_24155) | 46540..47106 | - | 567 | WP_000399780 | type IV conjugative transfer system protein TraE | traE |
| BTN71_RS24315 (BTN71_24160) | 47128..47439 | - | 312 | WP_000012113 | type IV conjugative transfer system protein TraL | traL |
| BTN71_RS24320 (BTN71_24165) | 47454..47813 | - | 360 | WP_001098992 | type IV conjugative transfer system pilin TraA | - |
| BTN71_RS24325 (BTN71_24170) | 47846..48073 | - | 228 | WP_000089263 | conjugal transfer relaxosome protein TraY | - |
| BTN71_RS24330 (BTN71_24175) | 48210..48881 | - | 672 | WP_000283561 | conjugal transfer transcriptional regulator TraJ | - |
| BTN71_RS24335 (BTN71_24180) | 49075..49458 | - | 384 | WP_001354030 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| BTN71_RS24345 (BTN71_24185) | 49793..50383 | + | 591 | WP_000252683 | transglycosylase SLT domain-containing protein | virB1 |
| BTN71_RS24350 (BTN71_24190) | 50680..51501 | - | 822 | WP_001234445 | DUF932 domain-containing protein | - |
| BTN71_RS24355 (BTN71_24195) | 51612..51908 | - | 297 | WP_001272251 | hypothetical protein | - |
| BTN71_RS25030 | 52208..52504 | + | 297 | Protein_68 | hypothetical protein | - |
| BTN71_RS24930 (BTN71_24215) | 52820..52945 | - | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| BTN71_RS25295 | 52887..53036 | - | 150 | Protein_70 | plasmid maintenance protein Mok | - |
| BTN71_RS24380 (BTN71_24225) | 53215..53977 | - | 763 | Protein_71 | plasmid SOS inhibition protein A | - |
| BTN71_RS24385 (BTN71_24230) | 53974..54408 | - | 435 | WP_000845923 | conjugation system SOS inhibitor PsiB | - |
Region 2: 78210..88535
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BTN71_RS24530 (BTN71_24385) | 73753..74882 | + | 1130 | WP_085959879 | IS3-like element IS1133 family transposase | - |
| BTN71_RS24535 (BTN71_24390) | 75014..75817 | + | 804 | WP_001082319 | aminoglycoside O-phosphotransferase APH(3'')-Ib | - |
| BTN71_RS24540 (BTN71_24395) | 75817..76653 | + | 837 | WP_000480968 | aminoglycoside O-phosphotransferase APH(6)-Id | - |
| BTN71_RS24545 (BTN71_24405) | 76779..77090 | - | 312 | WP_001452736 | hypothetical protein | - |
| BTN71_RS24550 (BTN71_24410) | 77126..77440 | - | 315 | WP_000734776 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| BTN71_RS24555 (BTN71_24415) | 77403..77780 | - | 378 | WP_000504251 | hypothetical protein | - |
| BTN71_RS24560 (BTN71_24420) | 77796..78101 | - | 306 | WP_000960954 | H-NS family nucleoid-associated regulatory protein | - |
| BTN71_RS24565 (BTN71_24425) | 78210..78944 | + | 735 | WP_000033783 | lytic transglycosylase domain-containing protein | virB1 |
| BTN71_RS24570 (BTN71_24430) | 78953..79234 | + | 282 | WP_000440698 | transcriptional repressor KorA | - |
| BTN71_RS24575 (BTN71_24435) | 79244..79537 | + | 294 | WP_000209137 | TrbC/VirB2 family protein | virB2 |
| BTN71_RS24580 (BTN71_24440) | 79587..79904 | + | 318 | WP_000496058 | VirB3 family type IV secretion system protein | virB3 |
| BTN71_RS24585 (BTN71_24445) | 79904..82504 | + | 2601 | WP_001200711 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| BTN71_RS24590 (BTN71_24450) | 82522..83235 | + | 714 | WP_000749362 | type IV secretion system protein | virB5 |
| BTN71_RS24595 (BTN71_24455) | 83243..83470 | + | 228 | WP_000734973 | IncN-type entry exclusion lipoprotein EexN | - |
| BTN71_RS24600 (BTN71_24460) | 83486..84526 | + | 1041 | WP_000886022 | type IV secretion system protein | virB6 |
| BTN71_RS25410 (BTN71_24465) | 84627..84755 | + | 129 | WP_071882546 | conjugal transfer protein TraN | - |
| BTN71_RS24610 (BTN71_24470) | 84745..85443 | + | 699 | WP_000646594 | type IV secretion system protein | virB8 |
| BTN71_RS24615 (BTN71_24475) | 85454..86338 | + | 885 | WP_000735067 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| BTN71_RS24620 (BTN71_24480) | 86338..87498 | + | 1161 | WP_000101711 | type IV secretion system protein VirB10 | virB10 |
| BTN71_RS24625 (BTN71_24485) | 87540..88535 | + | 996 | WP_000128596 | ATPase, T2SS/T4P/T4SS family | virB11 |
| BTN71_RS24630 (BTN71_24490) | 88535..89068 | + | 534 | WP_000731968 | phospholipase D family protein | - |
| BTN71_RS25415 | 89242..89637 | - | 396 | WP_306766596 | DUF6710 family protein | - |
| BTN71_RS24640 (BTN71_24500) | 89748..91499 | - | 1752 | Protein_120 | Tn3-like element TnAs1 family transposase | - |
| BTN71_RS24645 (BTN71_24505) | 91503..91895 | + | 393 | WP_001351729 | isochorismatase family cysteine hydrolase | - |
| BTN71_RS24650 (BTN71_24510) | 92033..92917 | + | 885 | WP_000058717 | EamA family transporter | - |
Host bacterium
| ID | 3340 | GenBank | NZ_CP018643 |
| Plasmid name | pSE74-1357 | Incompatibility group | IncN |
| Plasmid size | 118824 bp | Coordinate of oriT [Strand] | 111288..111388 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar Enteritidis strain 74-1357 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |