Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102874 |
| Name | oriT_pRsp8C3b |
| Organism | Rhizobium etli 8C-3 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP017243 (195436..195464 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRsp8C3b
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file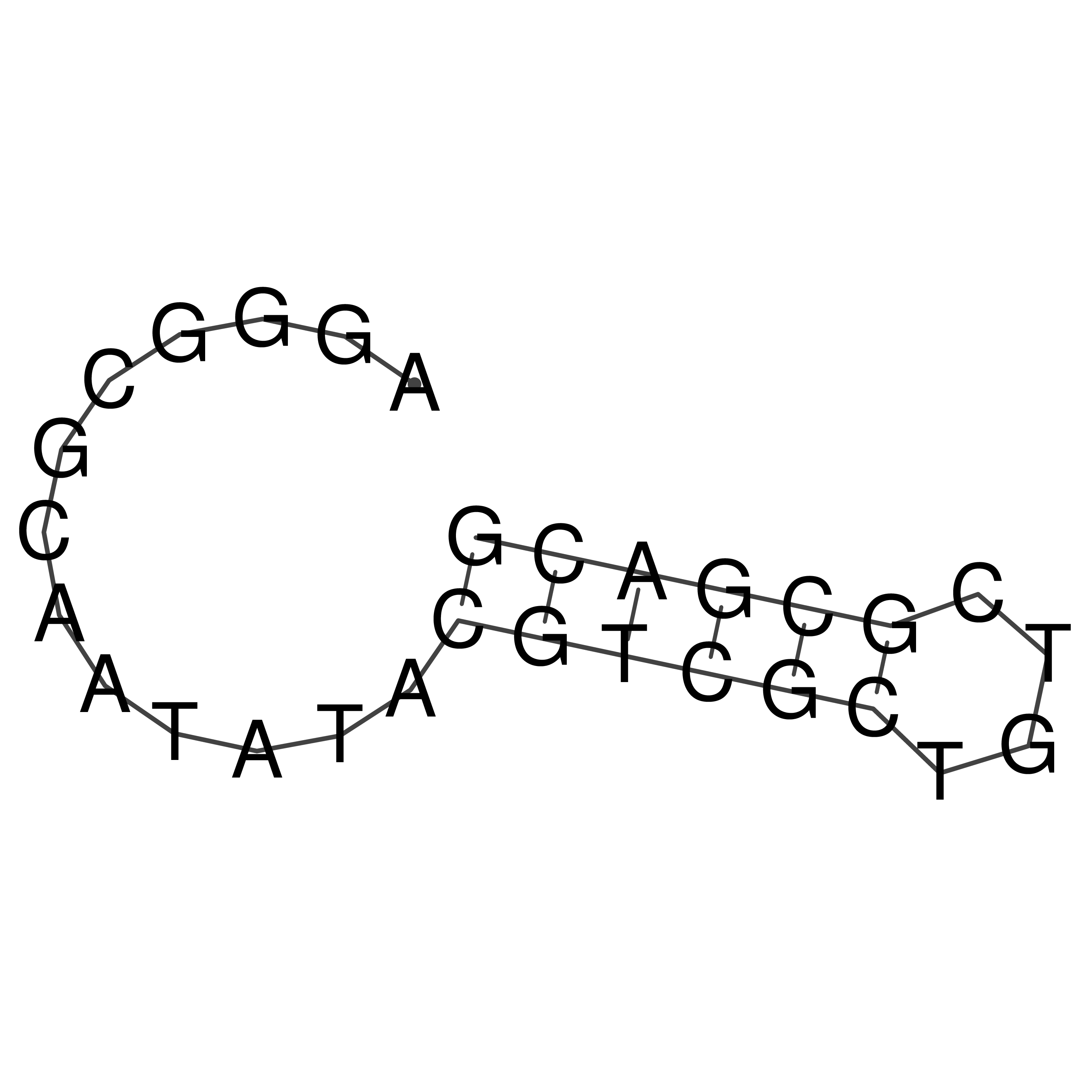
Relaxase
| ID | 2153 | GenBank | WP_018247162 |
| Name | traA_AM571_RS23495_pRsp8C3b |
UniProt ID | A0A1L5PBL4 |
| Length | 1552 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1552 a.a. Molecular weight: 171782.96 Da Isoelectric Point: 7.3778
>WP_018247162.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAGELMYEELALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENITLVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPATEEGFGAKKVPVLGEGGKPLRVVTPDRPNGKIVYKVWAGDKETMKAWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDERDIARALHRYVDDPTDFANIRARLMASDQLVILKPKEIEAETGKVSEPAVFTTREMLRIE
YDMAQSARVLSERRGFGVSERNVTVAIERVESIESGDPKTPFRLDAEQVDAVRHVTGDGGIAAIVGLAGA
GKSTLLAAARLAWEGEGHRVIGAALAGKAAEGLQDSSGIKSRTLASWELAWANGRDTLHRGDVLVIDEAG
MVASQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARNASRLFA
RGEVEKGLDAYTQQGHLVEAGTREETIDRIVVDWTEARKHAIGRSISEGRDGRLRGDELLVLAHTNDDVR
KLNEALREVMAGDNALGESRSFRTERGARKFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVTATG
DKRGDPLLSVLLDNGNKVVFGEDSYDNVDHGYAATIHKSQGSTVDRTFVLATGMMDRHLTYVSMTRHRDR
VDLYAAKEDFEPRPEWGRKPRVDHAAGVTGELVETGEAKFRPEDEDADDSPYADVRTDDGTAHRLWGVSL
PKALDDAGISDGDTITLRKDGVERVKVQIAIVDEETGQKRYEEREVDRNVWTARQIETAEARQERIERES
HRPDVFSRLVERLSRSGAKTTTLDFESEAGYRAQAQDFARRRGLDHLSLAAAEMEESLSRRWTWITAKRE
QVEKLWERASVALGFAIERERRFAYNEARTESHTIASASAGEPQYLIPPSTSFVRSVEADARLAQRSSPA
WTERETMLRPLLTKIYRDPDAALVALNALASDTGTEPRRLTADLAAAPDRLGRLRGSELIVDGGAALSER
KVAKAALEELLPLARAHATGFRRNAERFEMREQTRRSYMSLSIPALSERAMARLMEIEAVRNRDGDDAYK
SAFALAAEDRSVVQEIKAVSEALTARFGWSAFTAKADAVAERNMTERLPEDLTAGRREELARLFEAVKRF
AEAQHLAERQDRSKIVMAASVVRGQETENGPGKENVAVPPMLAAVIVFKTSVDDEARLRALSNPFYRQQR
GALANAATMIWRDPSGAVGKIEELLRKGFAADRIAAAVTNDPAAYGALRGSNRLLDRMLASGQERKEAMR
AVPEAAARLRALGAAHLNALDAERQAITDERRRMAVAIPALSKAAEEALAHLTVEVRKDSRKLSVSAASL
EQGIAREFAAVSRALDERFGRNALVRGDKDLVNRVPPAQRRAFEAMQERLKVLQQTVRLQSSEQIIAERR
QRAASRARGINL
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAGELMYEELALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENITLVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPATEEGFGAKKVPVLGEGGKPLRVVTPDRPNGKIVYKVWAGDKETMKAWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDERDIARALHRYVDDPTDFANIRARLMASDQLVILKPKEIEAETGKVSEPAVFTTREMLRIE
YDMAQSARVLSERRGFGVSERNVTVAIERVESIESGDPKTPFRLDAEQVDAVRHVTGDGGIAAIVGLAGA
GKSTLLAAARLAWEGEGHRVIGAALAGKAAEGLQDSSGIKSRTLASWELAWANGRDTLHRGDVLVIDEAG
MVASQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARNASRLFA
RGEVEKGLDAYTQQGHLVEAGTREETIDRIVVDWTEARKHAIGRSISEGRDGRLRGDELLVLAHTNDDVR
KLNEALREVMAGDNALGESRSFRTERGARKFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVTATG
DKRGDPLLSVLLDNGNKVVFGEDSYDNVDHGYAATIHKSQGSTVDRTFVLATGMMDRHLTYVSMTRHRDR
VDLYAAKEDFEPRPEWGRKPRVDHAAGVTGELVETGEAKFRPEDEDADDSPYADVRTDDGTAHRLWGVSL
PKALDDAGISDGDTITLRKDGVERVKVQIAIVDEETGQKRYEEREVDRNVWTARQIETAEARQERIERES
HRPDVFSRLVERLSRSGAKTTTLDFESEAGYRAQAQDFARRRGLDHLSLAAAEMEESLSRRWTWITAKRE
QVEKLWERASVALGFAIERERRFAYNEARTESHTIASASAGEPQYLIPPSTSFVRSVEADARLAQRSSPA
WTERETMLRPLLTKIYRDPDAALVALNALASDTGTEPRRLTADLAAAPDRLGRLRGSELIVDGGAALSER
KVAKAALEELLPLARAHATGFRRNAERFEMREQTRRSYMSLSIPALSERAMARLMEIEAVRNRDGDDAYK
SAFALAAEDRSVVQEIKAVSEALTARFGWSAFTAKADAVAERNMTERLPEDLTAGRREELARLFEAVKRF
AEAQHLAERQDRSKIVMAASVVRGQETENGPGKENVAVPPMLAAVIVFKTSVDDEARLRALSNPFYRQQR
GALANAATMIWRDPSGAVGKIEELLRKGFAADRIAAAVTNDPAAYGALRGSNRLLDRMLASGQERKEAMR
AVPEAAARLRALGAAHLNALDAERQAITDERRRMAVAIPALSKAAEEALAHLTVEVRKDSRKLSVSAASL
EQGIAREFAAVSRALDERFGRNALVRGDKDLVNRVPPAQRRAFEAMQERLKVLQQTVRLQSSEQIIAERR
QRAASRARGINL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1821 | GenBank | WP_004676100 |
| Name | traG_AM571_RS23480_pRsp8C3b |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70645.82 Da Isoelectric Point: 9.3989
>WP_004676100.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MGLRGKPHPSLLLVLVPIAVTTITIYIVGWRWPGLAAGMSGKIEHWFLRAAPVPPLLFGPLAGLLTVWVL
PLHRRRPVAMASLLCFLSVAAFYALREFGRLAPTVQARVVPWDRALSYLDMAAVIAAVAGFMAVAVSARI
SVVVPDEIKRARRGIFGDADWLPMAAAGKLFPPDGEIVVGERYRVDKEIIHALPFDVNDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPMICLDPSTEVAPMVVEHRTNALGREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSGSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNISASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNLSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAK
VNRFVKPVL
MGLRGKPHPSLLLVLVPIAVTTITIYIVGWRWPGLAAGMSGKIEHWFLRAAPVPPLLFGPLAGLLTVWVL
PLHRRRPVAMASLLCFLSVAAFYALREFGRLAPTVQARVVPWDRALSYLDMAAVIAAVAGFMAVAVSARI
SVVVPDEIKRARRGIFGDADWLPMAAAGKLFPPDGEIVVGERYRVDKEIIHALPFDVNDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPMICLDPSTEVAPMVVEHRTNALGREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSGSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNISASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNLSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAK
VNRFVKPVL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 174716..184303
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AM571_RS37605 (AM571_PB00170) | 171204..171429 | - | 226 | Protein_156 | chaperonin GroEL | - |
| AM571_RS23340 (AM571_PB00172) | 171724..172644 | + | 921 | WP_004673402 | zincin-like metallopeptidase domain-containing protein | - |
| AM571_RS23345 | 172755..172937 | - | 183 | WP_016737443 | hypothetical protein | - |
| AM571_RS23350 (AM571_PB00174) | 173706..174077 | - | 372 | WP_004673403 | hypothetical protein | - |
| AM571_RS23355 (AM571_PB00175) | 174176..174712 | + | 537 | WP_018446357 | hypothetical protein | - |
| AM571_RS23360 (AM571_PB00176) | 174716..175357 | + | 642 | WP_004673406 | transglycosylase SLT domain-containing protein | virB1 |
| AM571_RS23365 (AM571_PB00177) | 175372..175671 | + | 300 | WP_004673407 | TrbC/VirB2 family protein | virB2 |
| AM571_RS23370 (AM571_PB00178) | 175677..176015 | + | 339 | WP_018247156 | type IV secretion system protein VirB3 | virB3 |
| AM571_RS23375 (AM571_PB00179) | 176008..178374 | + | 2367 | WP_004673411 | VirB4 family type IV secretion system protein | virb4 |
| AM571_RS23380 (AM571_PB00180) | 178371..179072 | + | 702 | WP_004673414 | P-type DNA transfer protein VirB5 | virB5 |
| AM571_RS23385 (AM571_PB00181) | 179069..179302 | + | 234 | WP_004673415 | EexN family lipoprotein | - |
| AM571_RS23390 (AM571_PB00182) | 179305..180237 | + | 933 | WP_004673418 | type IV secretion system protein | virB6 |
| AM571_RS23395 (AM571_PB00183) | 180277..180558 | + | 282 | WP_004673420 | hypothetical protein | - |
| AM571_RS23400 (AM571_PB00184) | 180560..181231 | + | 672 | WP_004673422 | virB8 family protein | virB8 |
| AM571_RS23405 (AM571_PB00185) | 181228..182085 | + | 858 | WP_004673424 | P-type conjugative transfer protein VirB9 | virB9 |
| AM571_RS23410 (AM571_PB00186) | 182094..183266 | + | 1173 | WP_004673426 | type IV secretion system protein VirB10 | virB10 |
| AM571_RS23415 (AM571_PB00187) | 183275..184303 | + | 1029 | WP_004673428 | P-type DNA transfer ATPase VirB11 | virB11 |
| AM571_RS37610 | 184630..184923 | - | 294 | WP_004673430 | hypothetical protein | - |
| AM571_RS23425 (AM571_PB00189) | 184913..185359 | - | 447 | WP_004673432 | hypothetical protein | - |
| AM571_RS23430 (AM571_PB00191) | 185872..187062 | - | 1191 | WP_026188839 | DUF1173 domain-containing protein | - |
| AM571_RS37070 (AM571_PB00192) | 187662..187856 | - | 195 | WP_010011691 | hypothetical protein | - |
| AM571_RS23435 (AM571_PB00193) | 187958..188898 | + | 941 | Protein_177 | DUF2493 domain-containing protein | - |
| AM571_RS23440 (AM571_PB00195) | 188983..189249 | + | 267 | WP_004676093 | hypothetical protein | - |
Host bacterium
| ID | 3317 | GenBank | NZ_CP017243 |
| Plasmid name | pRsp8C3b | Incompatibility group | - |
| Plasmid size | 438379 bp | Coordinate of oriT [Strand] | 195436..195464 [+] |
| Host baterium | Rhizobium etli 8C-3 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | hsiC1/vipB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nifX, nifN, nifE, nifK, nifD, nifH, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, nifT, a9174_31165, nodD, nodJ, nodI, nodS, nodC, fixO, fixN, mLTONO_5203, nodA |
| Anti-CRISPR | - |