Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102860 |
| Name | oriT_pKPGJ-1a |
| Organism | Klebsiella variicola strain GJ1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP017283 (92220..92276 [+], 57 nt) |
| oriT length | 57 nt |
| IRs (inverted repeats) | 13..20, 23..30 (GCAAAATT..AATTTTGC) |
| Location of nic site | 39..40 |
| Conserved sequence flanking the nic site |
GGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 57 nt
>oriT_pKPGJ-1a
TAAAGTTAATCAGCAAAATTTTAATTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAG
TAAAGTTAATCAGCAAAATTTTAATTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file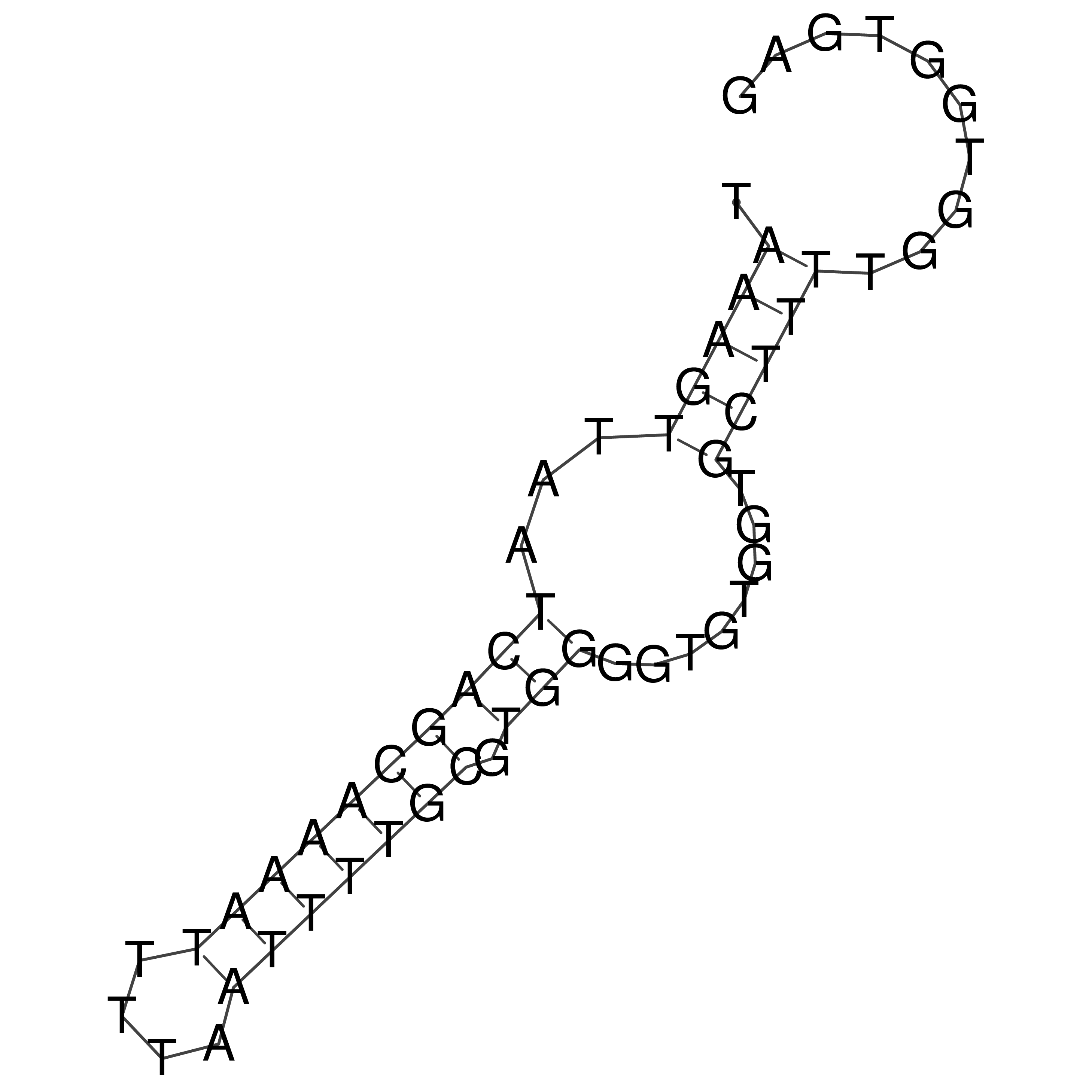
Relaxase
| ID | 2134 | GenBank | WP_077138950 |
| Name | traI_BBD63_RS01370_pKPGJ-1a |
UniProt ID | _ |
| Length | 1746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1746 a.a. Molecular weight: 189412.54 Da Isoelectric Point: 6.0094
>WP_077138950.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLFEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMQT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMTFTPGSTLIVDQGETLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGSREQAVLAGVVREALKAEKVLGEREVTIT
TLEPVWLDSRHQQVRDHYREGMVMERWSAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLKGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVGQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVQLYSAQDMQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQI
RSGALLSVPVSPGNGLQLLVSRQSYNAEKSILRHVLEGKEAVTPLMDKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERPDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIERVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQDAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVTLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLRPETTLAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGASLLARDMEEGVRLARENPQSGVVVRLEGEVRPWN
PGAITGGRVWADRLPDGTGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMERIKETPASPPTLPDAPEERRRDEAVSQVVRENVQRDRLQQMERETVRDLEREKTLGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLFEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMQT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMTFTPGSTLIVDQGETLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGSREQAVLAGVVREALKAEKVLGEREVTIT
TLEPVWLDSRHQQVRDHYREGMVMERWSAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLKGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVGQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVQLYSAQDMQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQI
RSGALLSVPVSPGNGLQLLVSRQSYNAEKSILRHVLEGKEAVTPLMDKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERPDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIERVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQDAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVTLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLRPETTLAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGASLLARDMEEGVRLARENPQSGVVVRLEGEVRPWN
PGAITGGRVWADRLPDGTGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMERIKETPASPPTLPDAPEERRRDEAVSQVVRENVQRDRLQQMERETVRDLEREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1797 | GenBank | WP_077138951 |
| Name | traD_BBD63_RS01375_pKPGJ-1a |
UniProt ID | _ |
| Length | 758 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 758 a.a. Molecular weight: 85004.73 Da Isoelectric Point: 5.1609
>WP_077138951.1 type IV conjugative transfer system coupling protein TraD [Klebsiella variicola]
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENAGQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQPQQPQQPQQPQQLAPAPRAADTASASASVAGAAGTGGVEPELKTKAEEA
EQLPPGISESGEVVDMAAWEAWQAEQDELSPQERQRREEVNINVHRAPEKDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENAGQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQPQQPQQPQQPQQLAPAPRAADTASASASVAGAAGTGGVEPELKTKAEEA
EQLPPGISESGEVVDMAAWEAWQAEQDELSPQERQRREEVNINVHRAPEKDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1798 | GenBank | WP_077138961 |
| Name | traC_BBD63_RS01445_pKPGJ-1a |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 98782.49 Da Isoelectric Point: 6.0216
>WP_077138961.1 MULTISPECIES: type IV secretion system protein TraC [Enterobacteriaceae]
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRNGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHAHEGKTIS
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRNGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHAHEGKTIS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 67713..92824
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BBD63_RS01375 (BBD63_26565) | 67713..69989 | - | 2277 | WP_077138951 | type IV conjugative transfer system coupling protein TraD | virb4 |
| BBD63_RS01380 (BBD63_26570) | 70123..70797 | - | 675 | WP_077138952 | tetratricopeptide repeat protein | - |
| BBD63_RS01385 (BBD63_26575) | 70872..71303 | - | 432 | WP_077138953 | entry exclusion protein | - |
| BBD63_RS01390 (BBD63_26580) | 71325..74147 | - | 2823 | WP_077138954 | conjugal transfer mating-pair stabilization protein TraG | traG |
| BBD63_RS01395 (BBD63_26585) | 74147..75481 | - | 1335 | WP_230199841 | conjugal transfer pilus assembly protein TraH | traH |
| BBD63_RS01400 (BBD63_26590) | 75520..76065 | - | 546 | WP_077138955 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| BBD63_RS01405 (BBD63_26595) | 76055..76303 | - | 249 | WP_008324126 | type-F conjugative transfer system pilin chaperone TraQ | - |
| BBD63_RS01410 (BBD63_26600) | 76320..77057 | - | 738 | WP_077138956 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| BBD63_RS01415 (BBD63_26605) | 77100..77288 | - | 189 | WP_077138957 | hypothetical protein | - |
| BBD63_RS01420 (BBD63_26610) | 77320..79200 | - | 1881 | WP_077138958 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| BBD63_RS01425 (BBD63_26615) | 79197..79826 | - | 630 | WP_077138959 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| BBD63_RS01430 (BBD63_26620) | 79843..80829 | - | 987 | WP_032637387 | conjugal transfer pilus assembly protein TraU | traU |
| BBD63_RS01435 (BBD63_26625) | 80832..81479 | - | 648 | WP_077138960 | type-F conjugative transfer system protein TraW | traW |
| BBD63_RS01440 (BBD63_26630) | 81479..81826 | - | 348 | WP_011201813 | type-F conjugative transfer system protein TrbI | - |
| BBD63_RS01445 (BBD63_26635) | 81823..84453 | - | 2631 | WP_077138961 | type IV secretion system protein TraC | virb4 |
| BBD63_RS30960 | 84446..85114 | - | 669 | WP_227516824 | conjugal transfer protein TraP | - |
| BBD63_RS30965 | 85104..85274 | - | 171 | WP_227516825 | Clp protease | - |
| BBD63_RS01460 | 85492..85677 | - | 186 | WP_016674194 | hypothetical protein | - |
| BBD63_RS01465 (BBD63_26650) | 85681..85905 | - | 225 | WP_016674195 | TraR/DksA C4-type zinc finger protein | - |
| BBD63_RS01470 (BBD63_26655) | 86025..86564 | - | 540 | WP_077138963 | type IV conjugative transfer system lipoprotein TraV | traV |
| BBD63_RS01475 (BBD63_26660) | 86586..87980 | - | 1395 | WP_060446827 | F-type conjugal transfer pilus assembly protein TraB | traB |
| BBD63_RS01480 (BBD63_26665) | 87967..88710 | - | 744 | WP_077138964 | type-F conjugative transfer system secretin TraK | traK |
| BBD63_RS01485 (BBD63_26670) | 88697..89263 | - | 567 | WP_077138965 | type IV conjugative transfer system protein TraE | traE |
| BBD63_RS01490 (BBD63_26675) | 89278..89583 | - | 306 | WP_071667439 | type IV conjugative transfer system protein TraL | traL |
| BBD63_RS01495 (BBD63_26680) | 89734..90084 | - | 351 | WP_077138966 | type IV conjugative transfer system pilin TraA | - |
| BBD63_RS30970 (BBD63_26685) | 90150..90365 | - | 216 | WP_321187714 | TraY domain-containing protein | - |
| BBD63_RS29565 | 91127..91336 | - | 210 | WP_145952642 | hypothetical protein | - |
| BBD63_RS01505 (BBD63_26690) | 91521..91916 | - | 396 | WP_040218546 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| BBD63_RS01510 (BBD63_26695) | 92348..92824 | + | 477 | WP_058673456 | transglycosylase SLT domain-containing protein | virB1 |
| BBD63_RS01515 (BBD63_26700) | 92870..93376 | - | 507 | Protein_109 | antirestriction protein | - |
| BBD63_RS01520 (BBD63_26705) | 93857..94687 | - | 831 | WP_077138967 | N-6 DNA methylase | - |
| BBD63_RS01525 (BBD63_26710) | 94730..94993 | - | 264 | WP_008324174 | hypothetical protein | - |
| BBD63_RS01530 (BBD63_26715) | 95079..95423 | - | 345 | WP_077138968 | hypothetical protein | - |
| BBD63_RS01535 (BBD63_26720) | 95470..95757 | - | 288 | WP_077138969 | hypothetical protein | - |
| BBD63_RS01540 (BBD63_26725) | 95818..96213 | - | 396 | WP_077138970 | ammonia monooxygenase | - |
| BBD63_RS29570 | 96636..97118 | - | 483 | WP_193791192 | conjugation system SOS inhibitor PsiB | - |
Host bacterium
| ID | 3303 | GenBank | NZ_CP017283 |
| Plasmid name | pKPGJ-1a | Incompatibility group | IncFII |
| Plasmid size | 108867 bp | Coordinate of oriT [Strand] | 92220..92276 [+] |
| Host baterium | Klebsiella variicola strain GJ1 |
Cargo genes
| Drug resistance gene | blaNDM-9, sul1, qacE, aadA2, dfrA12, aph(3')-Ia |
| Virulence gene | - |
| Metal resistance gene | merP, merT, merE, merD |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |