Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102827 |
| Name | oriT_pNY9_2 |
| Organism | Klebsiella pneumoniae strain NY9 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP015387 (26486..26534 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pNY9_2
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file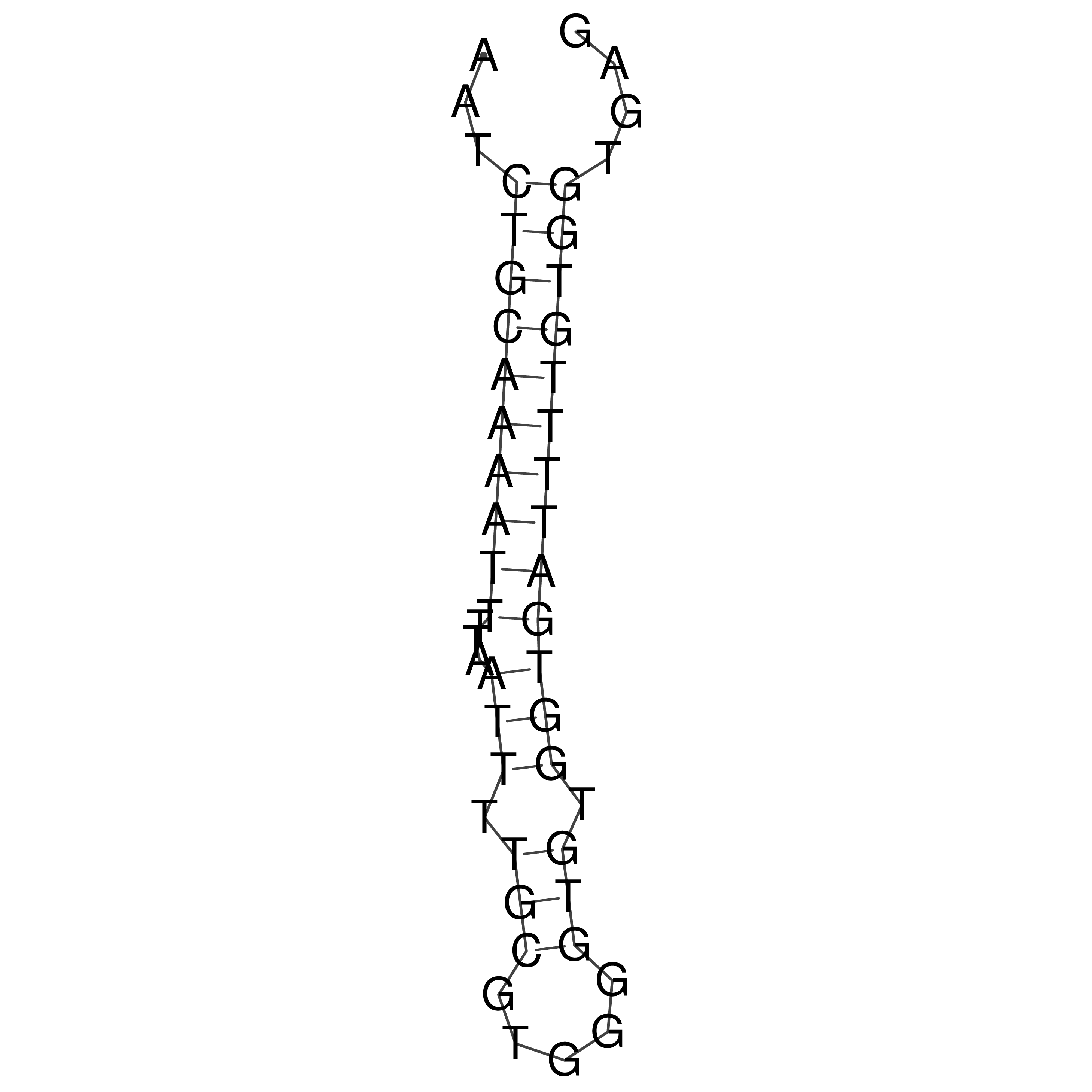
Relaxase
| ID | 2095 | GenBank | WP_022644926 |
| Name | traI_A6P56_RS29755_pNY9_2 |
UniProt ID | W8RBA8 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190574.57 Da Isoelectric Point: 6.3191
>WP_022644926.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLPPIAPGQPFRLTQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLPPIAPGQPFRLTQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 842 | GenBank | WP_022644939 |
| Name | WP_022644939_pNY9_2 |
UniProt ID | W8QGP1 |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 15048.16 Da Isoelectric Point: 4.5248
>WP_022644939.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Enterobacteriaceae]
MAKVQAYVSDEVAEKINAIVEKRRVEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNKAL
LEYVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMLRFFPDEDNEVEE
MAKVQAYVSDEVAEKINAIVEKRRVEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNKAL
LEYVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMLRFFPDEDNEVEE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | W8QGP1 |
T4CP
| ID | 1755 | GenBank | WP_032072078 |
| Name | traD_A6P56_RS29750_pNY9_2 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85822.78 Da Isoelectric Point: 4.9615
>WP_032072078.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 28911..54856
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| A6P56_RS29555 (A6P56_28190) | 24134..24481 | + | 348 | WP_022644944 | hypothetical protein | - |
| A6P56_RS29560 (A6P56_28195) | 24574..24720 | + | 147 | WP_022644943 | hypothetical protein | - |
| A6P56_RS34740 | 24768..25004 | + | 237 | Protein_34 | SAM-dependent DNA methyltransferase | - |
| A6P56_RS29570 (A6P56_28205) | 25353..25895 | + | 543 | WP_022644941 | antirestriction protein | - |
| A6P56_RS29575 (A6P56_28210) | 25919..26341 | - | 423 | WP_072205880 | transglycosylase SLT domain-containing protein | - |
| A6P56_RS29580 (A6P56_28215) | 26847..27239 | + | 393 | WP_022644939 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| A6P56_RS29585 (A6P56_28220) | 27474..28175 | + | 702 | WP_022644938 | hypothetical protein | - |
| A6P56_RS29590 | 28260..28460 | + | 201 | WP_032072063 | TraY domain-containing protein | - |
| A6P56_RS29595 (A6P56_28225) | 28529..28897 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| A6P56_RS29600 (A6P56_28230) | 28911..29216 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| A6P56_RS29605 (A6P56_28235) | 29236..29802 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| A6P56_RS29610 (A6P56_28240) | 29789..30529 | + | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| A6P56_RS29615 (A6P56_28245) | 30529..31953 | + | 1425 | WP_022644937 | F-type conjugal transfer pilus assembly protein TraB | traB |
| A6P56_RS29620 (A6P56_28250) | 32067..32651 | + | 585 | WP_004161368 | type IV conjugative transfer system lipoprotein TraV | traV |
| A6P56_RS29625 (A6P56_28255) | 32783..33193 | + | 411 | WP_004152499 | hypothetical protein | - |
| A6P56_RS29630 | 33299..33517 | + | 219 | WP_004152501 | hypothetical protein | - |
| A6P56_RS29635 (A6P56_28260) | 33518..33829 | + | 312 | WP_004152502 | hypothetical protein | - |
| A6P56_RS29640 (A6P56_28265) | 33896..34300 | + | 405 | WP_004152503 | hypothetical protein | - |
| A6P56_RS34420 | 34343..34732 | + | 390 | WP_004153076 | hypothetical protein | - |
| A6P56_RS29650 (A6P56_28275) | 34740..35138 | + | 399 | WP_011977783 | hypothetical protein | - |
| A6P56_RS29655 (A6P56_28280) | 35210..37849 | + | 2640 | WP_022644936 | type IV secretion system protein TraC | virb4 |
| A6P56_RS29660 (A6P56_28285) | 37849..38241 | + | 393 | WP_022644935 | type-F conjugative transfer system protein TrbI | - |
| A6P56_RS29665 (A6P56_28290) | 38238..38873 | + | 636 | WP_020325113 | type-F conjugative transfer system protein TraW | traW |
| A6P56_RS29670 (A6P56_28295) | 38908..39309 | + | 402 | WP_022644934 | hypothetical protein | - |
| A6P56_RS29675 (A6P56_28300) | 39306..40295 | + | 990 | WP_015632509 | conjugal transfer pilus assembly protein TraU | traU |
| A6P56_RS29680 (A6P56_28305) | 40308..40946 | + | 639 | WP_022644933 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| A6P56_RS29685 (A6P56_28310) | 41005..42960 | + | 1956 | WP_022644932 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| A6P56_RS29690 (A6P56_28315) | 42992..43246 | + | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| A6P56_RS29695 (A6P56_28320) | 43224..43472 | + | 249 | WP_022644931 | hypothetical protein | - |
| A6P56_RS29700 (A6P56_28325) | 43485..43811 | + | 327 | WP_004152676 | hypothetical protein | - |
| A6P56_RS29705 (A6P56_28330) | 43832..44584 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| A6P56_RS29710 (A6P56_28335) | 44595..44834 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| A6P56_RS29715 (A6P56_28340) | 44806..45363 | + | 558 | WP_022644930 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| A6P56_RS29720 (A6P56_28345) | 45409..45852 | + | 444 | WP_004194305 | F-type conjugal transfer protein TrbF | - |
| A6P56_RS29725 (A6P56_28350) | 45830..47209 | + | 1380 | WP_075995129 | conjugal transfer pilus assembly protein TraH | traH |
| A6P56_RS29730 (A6P56_28355) | 47209..50058 | + | 2850 | WP_004152624 | conjugal transfer mating-pair stabilization protein TraG | traG |
| A6P56_RS29735 (A6P56_28360) | 50071..50619 | + | 549 | WP_004152623 | conjugal transfer entry exclusion protein TraS | - |
| A6P56_RS29740 (A6P56_28365) | 50803..51534 | + | 732 | WP_004152622 | conjugal transfer complement resistance protein TraT | - |
| A6P56_RS34160 | 51726..52415 | + | 690 | WP_022644928 | hypothetical protein | - |
| A6P56_RS29750 (A6P56_28375) | 52544..54856 | + | 2313 | WP_032072078 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 3270 | GenBank | NZ_CP015387 |
| Plasmid name | pNY9_2 | Incompatibility group | - |
| Plasmid size | 139933 bp | Coordinate of oriT [Strand] | 26486..26534 [-] |
| Host baterium | Klebsiella pneumoniae strain NY9 |
Cargo genes
| Drug resistance gene | blaKPC-3, aac(6')-Ib, ant(3'')-Ia, blaOXA-9, blaTEM-1A, aph(6)-Id, aph(3'')-Ib, sul2, dfrA14 |
| Virulence gene | - |
| Metal resistance gene | ncrY, ncrC, ncrB, ncrA |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |