Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102797 |
| Name | oriT_Vaf10|unnamed4 |
| Organism | Rhizobium leguminosarum strain Vaf10 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP016292 (111666..111708 [-], 43 nt) |
| oriT length | 43 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 43 nt
>oriT_Vaf10|unnamed4
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTGC
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file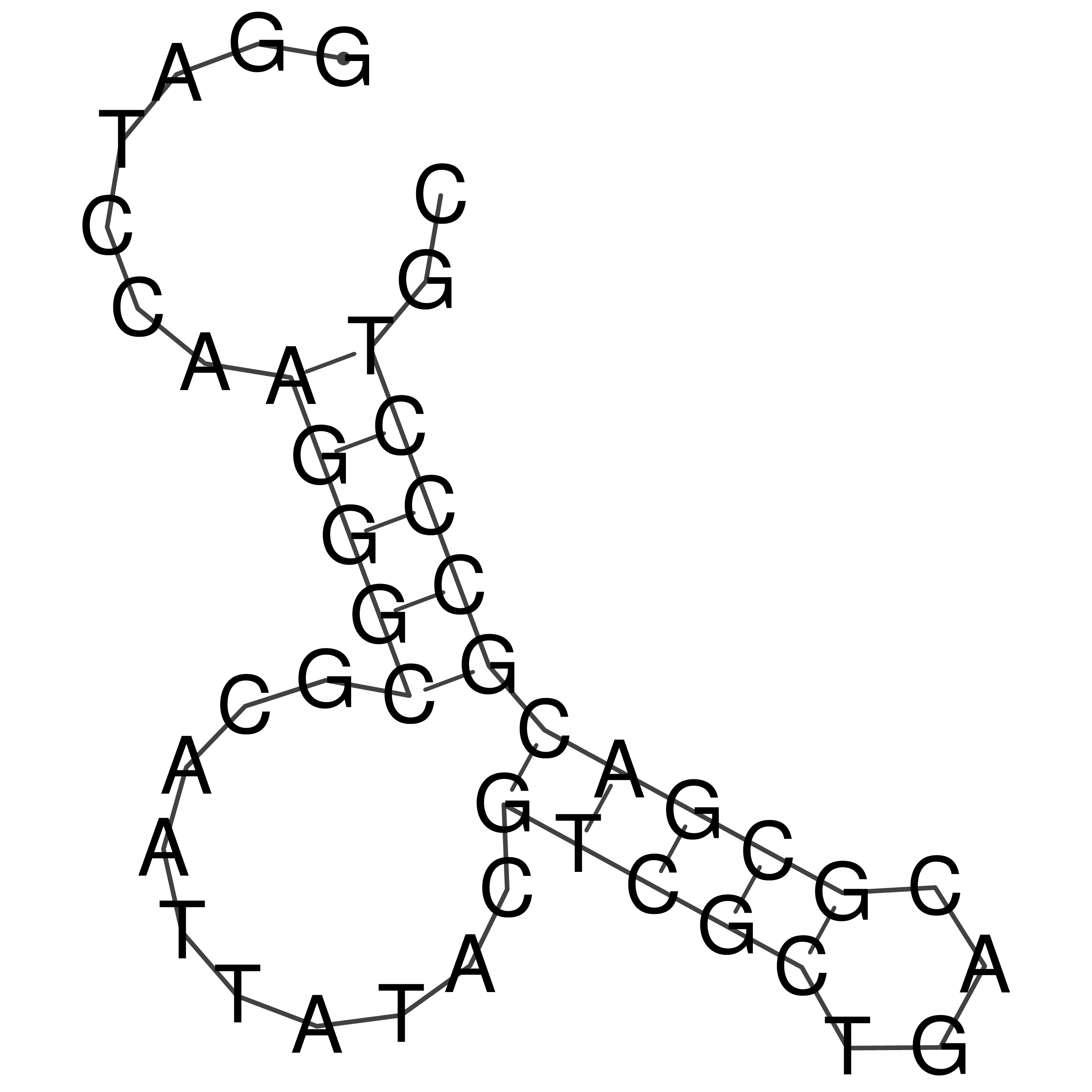
Relaxase
| ID | 2071 | GenBank | WP_065284726 |
| Name | traA_BA011_RS38865_Vaf10|unnamed4 |
UniProt ID | A0A1B1CP89 |
| Length | 1193 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1193 a.a. Molecular weight: 131490.11 Da Isoelectric Point: 8.8551
>WP_065284726.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYSRKQGLLHEEFVIPEDAPDWLRSMLADR
SVSGASEAFWNKVEAFEKRSDAQLAKDVTIALPIELSADQNIALVRDFVERHITAKGVVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGAKKVAVLGPDGNPVRNDAGKIVYELWAGSADDFNAFRDGWFACQNRHLALAG
FDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKGDSGEEKVALERLELQEERRAENARRIQRNPEIVVDL
ITREKSVFDERDIAKVLYRYIDDAALFQNLMARLLQSPQTLRLDRERMDLVTGVRAPAKYTTRELIRLEA
QMANQAIWLSQRSSHRVNRTVLSGIVSRHDRLSDEQKTAIEHVAGPERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAADGLEKEAGIASRTLSAWELRWDQERDRLDEKSVFVLDEAGMVSSRQMARFVE
AVTLSGAKLVLVGDPEQLQPIEAGAAFRAIAERIGYAELETIYRQREQWMRDASLDLARGNVSAALDAYA
QRDLVRTGWTRDEAITALIAEWDHEYDPAKSTLILAHRRVDVRLLNEMARSKLVERGLIEAGHAFKTEDG
TRQFAAGDQIVFLKNEGSLGVKNGMLALVVDAQPGRIVAEIGNGEDRRRVVVEQRFYANVDHGYATTVHK
SQGATVDRVKVLASSTLDRHLSYVAMTRHRETAELYVGLEEFAQRRGGVLIAHGEAPYEHKPGNRDSYYV
TLGFANGQERTVWGVDLARAMDASDSRIGDRIGLKHVGSQRVTLPNGTEVDRNSWKVVPIQELAMARLHE
RLSRAGSKETTLDYQDASHYRAALRFAEARGLHLMNVARTIAHDQLQWTVRQSSKLAELGARLVAVAAKL
GLGGAKSTVSTTSVIKEAKPMVFGTTTFPRSIGQAVEDKLSADPGLKASWQEVSARFHHVFADPQAAFKA
VNVDAMLANGTVAATTIVRIAEQPESFGALKGKTGLFAGSAEKQARDTALVNAPALARDLQGFIAKRAAA
ARRYEDEERAVRAQLSLDIPALSASGKQVLERVRDAIDRNDIPAGLEFALADKMVKAELEGFAKAVSERF
GERTFLPLAAKTADGKAFEVASAGMQPAQKNELRSAWDTIRTVQQLAAHERTAVALKQAEAIRQTQTKGL
SLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYSRKQGLLHEEFVIPEDAPDWLRSMLADR
SVSGASEAFWNKVEAFEKRSDAQLAKDVTIALPIELSADQNIALVRDFVERHITAKGVVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGAKKVAVLGPDGNPVRNDAGKIVYELWAGSADDFNAFRDGWFACQNRHLALAG
FDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKGDSGEEKVALERLELQEERRAENARRIQRNPEIVVDL
ITREKSVFDERDIAKVLYRYIDDAALFQNLMARLLQSPQTLRLDRERMDLVTGVRAPAKYTTRELIRLEA
QMANQAIWLSQRSSHRVNRTVLSGIVSRHDRLSDEQKTAIEHVAGPERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAADGLEKEAGIASRTLSAWELRWDQERDRLDEKSVFVLDEAGMVSSRQMARFVE
AVTLSGAKLVLVGDPEQLQPIEAGAAFRAIAERIGYAELETIYRQREQWMRDASLDLARGNVSAALDAYA
QRDLVRTGWTRDEAITALIAEWDHEYDPAKSTLILAHRRVDVRLLNEMARSKLVERGLIEAGHAFKTEDG
TRQFAAGDQIVFLKNEGSLGVKNGMLALVVDAQPGRIVAEIGNGEDRRRVVVEQRFYANVDHGYATTVHK
SQGATVDRVKVLASSTLDRHLSYVAMTRHRETAELYVGLEEFAQRRGGVLIAHGEAPYEHKPGNRDSYYV
TLGFANGQERTVWGVDLARAMDASDSRIGDRIGLKHVGSQRVTLPNGTEVDRNSWKVVPIQELAMARLHE
RLSRAGSKETTLDYQDASHYRAALRFAEARGLHLMNVARTIAHDQLQWTVRQSSKLAELGARLVAVAAKL
GLGGAKSTVSTTSVIKEAKPMVFGTTTFPRSIGQAVEDKLSADPGLKASWQEVSARFHHVFADPQAAFKA
VNVDAMLANGTVAATTIVRIAEQPESFGALKGKTGLFAGSAEKQARDTALVNAPALARDLQGFIAKRAAA
ARRYEDEERAVRAQLSLDIPALSASGKQVLERVRDAIDRNDIPAGLEFALADKMVKAELEGFAKAVSERF
GERTFLPLAAKTADGKAFEVASAGMQPAQKNELRSAWDTIRTVQQLAAHERTAVALKQAEAIRQTQTKGL
SLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1B1CP89 |
T4CP
| ID | 1731 | GenBank | WP_028744195 |
| Name | t4cp2_BA011_RS38770_Vaf10|unnamed4 |
UniProt ID | _ |
| Length | 811 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 811 a.a. Molecular weight: 90925.37 Da Isoelectric Point: 5.8208
>WP_028744195.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium/Agrobacterium group]
MVALRTFRSTGPSFADLVPYAGLVDNGILLLKDGSLMAGWYFAGPDSESATDFERNELSRQINSILSRLG
TGWMIQVEAVRVPTTEYPSAERSHFPDAVTLLIDHERRRHFGQERGHFESRHALILTYRPPERRRSGLTR
YIYSDNESRSAKYADTALDAFRRSIREIEQYLGNVLSVQRMQTREVSERDGARVARYDELFQFIRFAITG
ENHPVRLPEIPMYLDWLATAELEHGLTPRVEGRFLGVIAIDGLPAESWPGILNSLNLMPLTYRWSSRFIF
LDAEEAKQRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEDAIAEASSQLVAYGYYTPVIVL
FDESRSALQEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLVPLNSVWS
GQPFAPCPFYPPDAPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFCRYEKAQVFA
FDKGNSMLTLTLGVGGDHYEIGGESGEGASLAFCPLSELSTDADRAWASEWIETLVSLQGVTIAPDHRNA
ISRQIGLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGLLLDAEEDGLTLGRFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVILATQSISDAERS
GIIDVLKESCPTKICLPNGAARETGTREFYERIGFNARQIEIVANAIPKREYYVTSPDGRRLFDMSLGPV
TLSFVGASGKADLARIRALSSTHGLEWPGQWLIERGINRNE
MVALRTFRSTGPSFADLVPYAGLVDNGILLLKDGSLMAGWYFAGPDSESATDFERNELSRQINSILSRLG
TGWMIQVEAVRVPTTEYPSAERSHFPDAVTLLIDHERRRHFGQERGHFESRHALILTYRPPERRRSGLTR
YIYSDNESRSAKYADTALDAFRRSIREIEQYLGNVLSVQRMQTREVSERDGARVARYDELFQFIRFAITG
ENHPVRLPEIPMYLDWLATAELEHGLTPRVEGRFLGVIAIDGLPAESWPGILNSLNLMPLTYRWSSRFIF
LDAEEAKQRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEDAIAEASSQLVAYGYYTPVIVL
FDESRSALQEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLVPLNSVWS
GQPFAPCPFYPPDAPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFCRYEKAQVFA
FDKGNSMLTLTLGVGGDHYEIGGESGEGASLAFCPLSELSTDADRAWASEWIETLVSLQGVTIAPDHRNA
ISRQIGLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGLLLDAEEDGLTLGRFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVILATQSISDAERS
GIIDVLKESCPTKICLPNGAARETGTREFYERIGFNARQIEIVANAIPKREYYVTSPDGRRLFDMSLGPV
TLSFVGASGKADLARIRALSSTHGLEWPGQWLIERGINRNE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1732 | GenBank | WP_065284729 |
| Name | traG_BA011_RS38880_Vaf10|unnamed4 |
UniProt ID | _ |
| Length | 656 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 656 a.a. Molecular weight: 70653.58 Da Isoelectric Point: 9.4607
>WP_065284729.1 Ti-type conjugative transfer system protein TraG [Rhizobium leguminosarum]
MTPKRILVAGIPAVAMLAIALTVPGIERWLSAFGTTDQAKLVLGRIGLALPYALAGACGVMFLFGTKGSI
NVKTSGWSVAAGGLVVIIVAALREGSRLLAFAGRVPADRALLSYADPSTIAGAAVTLLSVFFALRVARTG
NAAFARSEPRRIRGKRALHGEADWMTMQGSEKLFPETGGIVIGERYRVDRDDPAAAGFRAGEPTTWGRGG
SAPLLCFDGSFGSSHGIVFAGSGGYKTTSVTIPTALKWGGSLVVLDPSNEVAPMVMEHRRKAGRRVIVLD
PKNAGSGFNALDWIGRHGGTKEEDIAAVASWIMSDSGRATGVRDDFFRASGLQLLTAMIADVCLSGHTDA
KHQTLRQVRANLSEPEPKLRARLQEIYDSSASDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYTNYA
ALVSGSTFSTDALADGETDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGEVKGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRESYGGRDAASKWFESVSWISFAAINDPDTADYISKRCGMTT
VEIDQVSRSFQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNPPLRCGRAIWFRRDDMKACVG
TNRFHKIGGPAGSTSDRADAECNEQG
MTPKRILVAGIPAVAMLAIALTVPGIERWLSAFGTTDQAKLVLGRIGLALPYALAGACGVMFLFGTKGSI
NVKTSGWSVAAGGLVVIIVAALREGSRLLAFAGRVPADRALLSYADPSTIAGAAVTLLSVFFALRVARTG
NAAFARSEPRRIRGKRALHGEADWMTMQGSEKLFPETGGIVIGERYRVDRDDPAAAGFRAGEPTTWGRGG
SAPLLCFDGSFGSSHGIVFAGSGGYKTTSVTIPTALKWGGSLVVLDPSNEVAPMVMEHRRKAGRRVIVLD
PKNAGSGFNALDWIGRHGGTKEEDIAAVASWIMSDSGRATGVRDDFFRASGLQLLTAMIADVCLSGHTDA
KHQTLRQVRANLSEPEPKLRARLQEIYDSSASDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYTNYA
ALVSGSTFSTDALADGETDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGEVKGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRESYGGRDAASKWFESVSWISFAAINDPDTADYISKRCGMTT
VEIDQVSRSFQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNPPLRCGRAIWFRRDDMKACVG
TNRFHKIGGPAGSTSDRADAECNEQG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 89802..106274
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BA011_RS38740 (BA011_36320) | 85756..87000 | - | 1245 | WP_065284716 | plasmid replication protein RepC | - |
| BA011_RS38745 (BA011_36325) | 87158..88150 | - | 993 | WP_028744193 | plasmid partitioning protein RepB | - |
| BA011_RS38750 (BA011_36330) | 88232..89422 | - | 1191 | WP_018009855 | plasmid partitioning protein RepA | - |
| BA011_RS38755 (BA011_36335) | 89802..90767 | + | 966 | WP_065284717 | P-type conjugative transfer ATPase TrbB | virB11 |
| BA011_RS38760 (BA011_36340) | 90757..91149 | + | 393 | WP_011654762 | TrbC/VirB2 family protein | virB2 |
| BA011_RS38765 (BA011_36345) | 91142..91441 | + | 300 | WP_011654761 | conjugal transfer protein TrbD | virB3 |
| BA011_RS38770 (BA011_36350) | 91452..93887 | + | 2436 | WP_028744195 | conjugal transfer protein TrbE | virb4 |
| BA011_RS38775 (BA011_36355) | 93880..94689 | + | 810 | WP_011654759 | P-type conjugative transfer protein TrbJ | virB5 |
| BA011_RS38780 (BA011_36360) | 94686..94886 | + | 201 | WP_028744197 | entry exclusion protein TrbK | - |
| BA011_RS38785 (BA011_36365) | 94880..96061 | + | 1182 | WP_065284718 | P-type conjugative transfer protein TrbL | virB6 |
| BA011_RS43795 | 96083..96229 | + | 147 | Protein_108 | conjugal transfer protein TrbF | - |
| BA011_RS38800 (BA011_36380) | 97471..97989 | + | 519 | Protein_110 | VirB8/TrbF family protein | - |
| BA011_RS38805 (BA011_36385) | 98041..98835 | + | 795 | WP_237352882 | P-type conjugative transfer protein TrbG | virB9 |
| BA011_RS38810 (BA011_36390) | 98839..99285 | + | 447 | WP_028744199 | conjugal transfer protein TrbH | - |
| BA011_RS38815 (BA011_36395) | 99301..100602 | + | 1302 | WP_065284720 | IncP-type conjugal transfer protein TrbI | virB10 |
| BA011_RS38820 (BA011_36400) | 100803..101102 | + | 300 | WP_027690597 | transcriptional repressor TraM | - |
| BA011_RS38825 (BA011_36405) | 101103..101807 | - | 705 | WP_027690598 | autoinducer binding domain-containing protein | - |
| BA011_RS38830 (BA011_36410) | 102090..103706 | + | 1617 | WP_065284721 | hypothetical protein | - |
| BA011_RS38835 (BA011_36415) | 103696..104691 | + | 996 | WP_065284722 | dienelactone hydrolase-related enzyme | - |
| BA011_RS38840 (BA011_36420) | 104840..105019 | + | 180 | WP_018010295 | hypothetical protein | - |
| BA011_RS38845 (BA011_36425) | 105016..105633 | + | 618 | WP_065284723 | hypothetical protein | - |
| BA011_RS38850 (BA011_36430) | 105657..106274 | - | 618 | WP_018010297 | TraH family protein | virB1 |
| BA011_RS38855 (BA011_36435) | 106290..107456 | - | 1167 | WP_065284724 | conjugal transfer protein TraB | - |
| BA011_RS38860 (BA011_36440) | 107446..108012 | - | 567 | WP_065284725 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 3240 | GenBank | NZ_CP016292 |
| Plasmid name | Vaf10|unnamed4 | Incompatibility group | - |
| Plasmid size | 309378 bp | Coordinate of oriT [Strand] | 111666..111708 [-] |
| Host baterium | Rhizobium leguminosarum strain Vaf10 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD |
| Anti-CRISPR | - |