Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102796 |
| Name | oriT_Vaf10|unnamed3 |
| Organism | Rhizobium leguminosarum strain Vaf10 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP016290 (256508..256541 [-], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_Vaf10|unnamed3
TCCAAGGGCGCAATTATACGTCGCCGGCGCGACG
TCCAAGGGCGCAATTATACGTCGCCGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file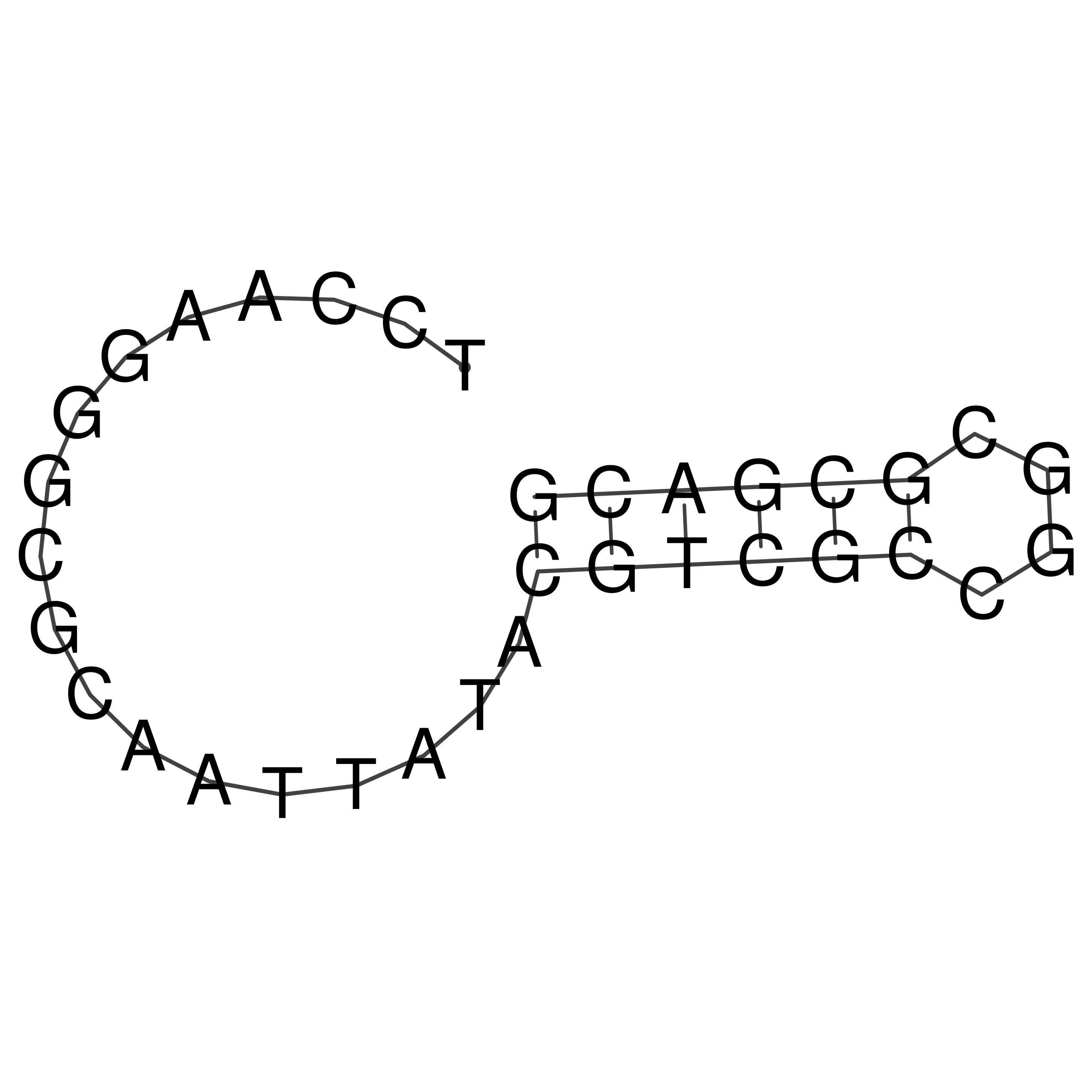
Relaxase
| ID | 2070 | GenBank | WP_065284310 |
| Name | traA_BA011_RS35695_Vaf10|unnamed3 |
UniProt ID | A0A1B1CMV8 |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 123224.57 Da Isoelectric Point: 9.9668
>WP_065284310.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAVPHFSVSVVGRGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADSPEWLRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLARDVTIALPIELTATQNIALVRDFVERHITSKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVLLFQSLMVRILRSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHSHLSYEQRTAIEHVAGASRIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNQGRNQLDHKTVFVLDEAGMVSSRQM
AVLVETVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDTYTAHGRMIGLRLKDEAVESLIAAWDRDYEPSKTSLILAHLRRDVRMLNDMARAKLIERGVVADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVLEAAPGRIVVEVGDGEHRRQVMIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYFGSRSFAKSGGLVPILSRRNAKETTLDYE
KASFYGQALRFAEARGLHLMNVARTIARDRLEWTVRQKQKLADLGARLAAIGAALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLERAVEDKLAADPGLKKQWEDVSTRFQLVYAKPEAAFKAINVDTMLKDQSVAQS
TLARIAEEPESFGALKGKTGLLASRGDKQDREKALANVPALARNLERYLRERAEAELKHQTEERAVRLKV
SVDIPALSSAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTSG
KTFETVTSGMTAGQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETLRQTKNQGLSLK
MAVPHFSVSVVGRGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADSPEWLRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLARDVTIALPIELTATQNIALVRDFVERHITSKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVLLFQSLMVRILRSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHSHLSYEQRTAIEHVAGASRIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNQGRNQLDHKTVFVLDEAGMVSSRQM
AVLVETVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDTYTAHGRMIGLRLKDEAVESLIAAWDRDYEPSKTSLILAHLRRDVRMLNDMARAKLIERGVVADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVLEAAPGRIVVEVGDGEHRRQVMIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYFGSRSFAKSGGLVPILSRRNAKETTLDYE
KASFYGQALRFAEARGLHLMNVARTIARDRLEWTVRQKQKLADLGARLAAIGAALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLERAVEDKLAADPGLKKQWEDVSTRFQLVYAKPEAAFKAINVDTMLKDQSVAQS
TLARIAEEPESFGALKGKTGLLASRGDKQDREKALANVPALARNLERYLRERAEAELKHQTEERAVRLKV
SVDIPALSSAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTSG
KTFETVTSGMTAGQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETLRQTKNQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1B1CMV8 |
T4CP
| ID | 1729 | GenBank | WP_065284294 |
| Name | t4cp2_BA011_RS35615_Vaf10|unnamed3 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91708.64 Da Isoelectric Point: 6.1282
>WP_065284294.1 conjugal transfer protein TrbE [Rhizobium leguminosarum]
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQVNAVLSRLG
SSWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYADKESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETLEREGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVVVL
FDRDREGLLEKAEAIRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPFAPCPFYPPKSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVAVQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKFEHGRDWPIHWLETRGVHDAASLLK
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQVNAVLSRLG
SSWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYADKESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETLEREGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVVVL
FDRDREGLLEKAEAIRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPFAPCPFYPPKSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVAVQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKFEHGRDWPIHWLETRGVHDAASLLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1730 | GenBank | WP_065284311 |
| Name | traG_BA011_RS35710_Vaf10|unnamed3 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69411.41 Da Isoelectric Point: 9.1846
>WP_065284311.1 Ti-type conjugative transfer system protein TraG [Rhizobium leguminosarum]
MTINRIALAVMPAALMILVVIGMTGIEQWLSGFDKTEAARLAWGRTGIALPYVASAAIGVLLLFSSAGSI
NIKQAGWGVVAGSSGTILVAAVRETVRLSGFMKTVPADKTVLAYLDPATSIGASTALLGACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEIAPMVSAHRSGAGRDVFVLD
PRKPGIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTDE
HDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFATNDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNREGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTHSKQLAARPLIQPHEVLRIRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMIGDTPKSA
MTINRIALAVMPAALMILVVIGMTGIEQWLSGFDKTEAARLAWGRTGIALPYVASAAIGVLLLFSSAGSI
NIKQAGWGVVAGSSGTILVAAVRETVRLSGFMKTVPADKTVLAYLDPATSIGASTALLGACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEIAPMVSAHRSGAGRDVFVLD
PRKPGIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTDE
HDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFATNDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNREGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTHSKQLAARPLIQPHEVLRIRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMIGDTPKSA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 237478..251369
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BA011_RS35580 (BA011_34430) | 232804..234018 | - | 1215 | WP_065284288 | plasmid replication protein RepC | - |
| BA011_RS35585 (BA011_34435) | 234209..235246 | - | 1038 | WP_065284289 | plasmid partitioning protein RepB | - |
| BA011_RS35590 (BA011_34440) | 235243..236466 | - | 1224 | WP_065284460 | plasmid partitioning protein RepA | - |
| BA011_RS35595 (BA011_34445) | 236843..237481 | + | 639 | WP_065284290 | acyl-homoserine-lactone synthase | - |
| BA011_RS35600 (BA011_34450) | 237478..238455 | + | 978 | WP_065284291 | P-type conjugative transfer ATPase TrbB | virB11 |
| BA011_RS35605 (BA011_34455) | 238445..238828 | + | 384 | WP_065284292 | TrbC/VirB2 family protein | virB2 |
| BA011_RS35610 (BA011_34460) | 238821..239120 | + | 300 | WP_065284293 | conjugal transfer protein TrbD | virB3 |
| BA011_RS35615 (BA011_34465) | 239131..241581 | + | 2451 | WP_065284294 | conjugal transfer protein TrbE | virb4 |
| BA011_RS35620 (BA011_34470) | 241559..242356 | + | 798 | WP_065284295 | P-type conjugative transfer protein TrbJ | virB5 |
| BA011_RS35625 (BA011_34475) | 242350..242523 | + | 174 | WP_065284296 | entry exclusion protein TrbK | - |
| BA011_RS35630 (BA011_34480) | 242528..243715 | + | 1188 | WP_065284297 | P-type conjugative transfer protein TrbL | virB6 |
| BA011_RS35635 (BA011_34485) | 243738..244400 | + | 663 | WP_065284298 | conjugal transfer protein TrbF | virB8 |
| BA011_RS35640 (BA011_34490) | 244418..245230 | + | 813 | WP_065284299 | P-type conjugative transfer protein TrbG | virB9 |
| BA011_RS35645 (BA011_34495) | 245234..245674 | + | 441 | WP_065284300 | conjugal transfer protein TrbH | - |
| BA011_RS35650 (BA011_34500) | 245686..246984 | + | 1299 | WP_065284301 | IncP-type conjugal transfer protein TrbI | virB10 |
| BA011_RS35655 (BA011_34505) | 247296..248000 | + | 705 | WP_065284302 | autoinducer binding domain-containing protein | - |
| BA011_RS35660 (BA011_34510) | 248004..248294 | - | 291 | WP_065284303 | transcriptional repressor TraM | - |
| BA011_RS35665 (BA011_34515) | 248552..248800 | + | 249 | WP_065284304 | helix-turn-helix transcriptional regulator | - |
| BA011_RS35670 (BA011_34520) | 249039..249656 | + | 618 | WP_072642654 | type IV toxin-antitoxin system AbiEi family antitoxin domain-containing protein | - |
| BA011_RS35675 (BA011_34525) | 249649..250517 | + | 869 | Protein_257 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| BA011_RS35680 (BA011_34530) | 250752..251369 | - | 618 | WP_065284307 | TraH family protein | virB1 |
| BA011_RS35685 (BA011_34535) | 251386..252549 | - | 1164 | WP_065284308 | conjugal transfer protein TraB | - |
| BA011_RS35690 (BA011_34540) | 252539..253075 | - | 537 | WP_065284309 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 3239 | GenBank | NZ_CP016290 |
| Plasmid name | Vaf10|unnamed3 | Incompatibility group | - |
| Plasmid size | 557598 bp | Coordinate of oriT [Strand] | 256508..256541 [-] |
| Host baterium | Rhizobium leguminosarum strain Vaf10 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | hupE |
| Degradation gene | - |
| Symbiosis gene | nodM, nodE, nodF, nodD, nodA, nodC, nodI, nodJ, nifB, fixX, fixC, fixB, fixA, nifH, nifD, nifK, nifE, nifN, fixN, fixO, fixQ |
| Anti-CRISPR | - |