Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102785 |
| Name | oriT_pRspN731c |
| Organism | Rhizobium sp. N731 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013604 (211511..211539 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRspN731c
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file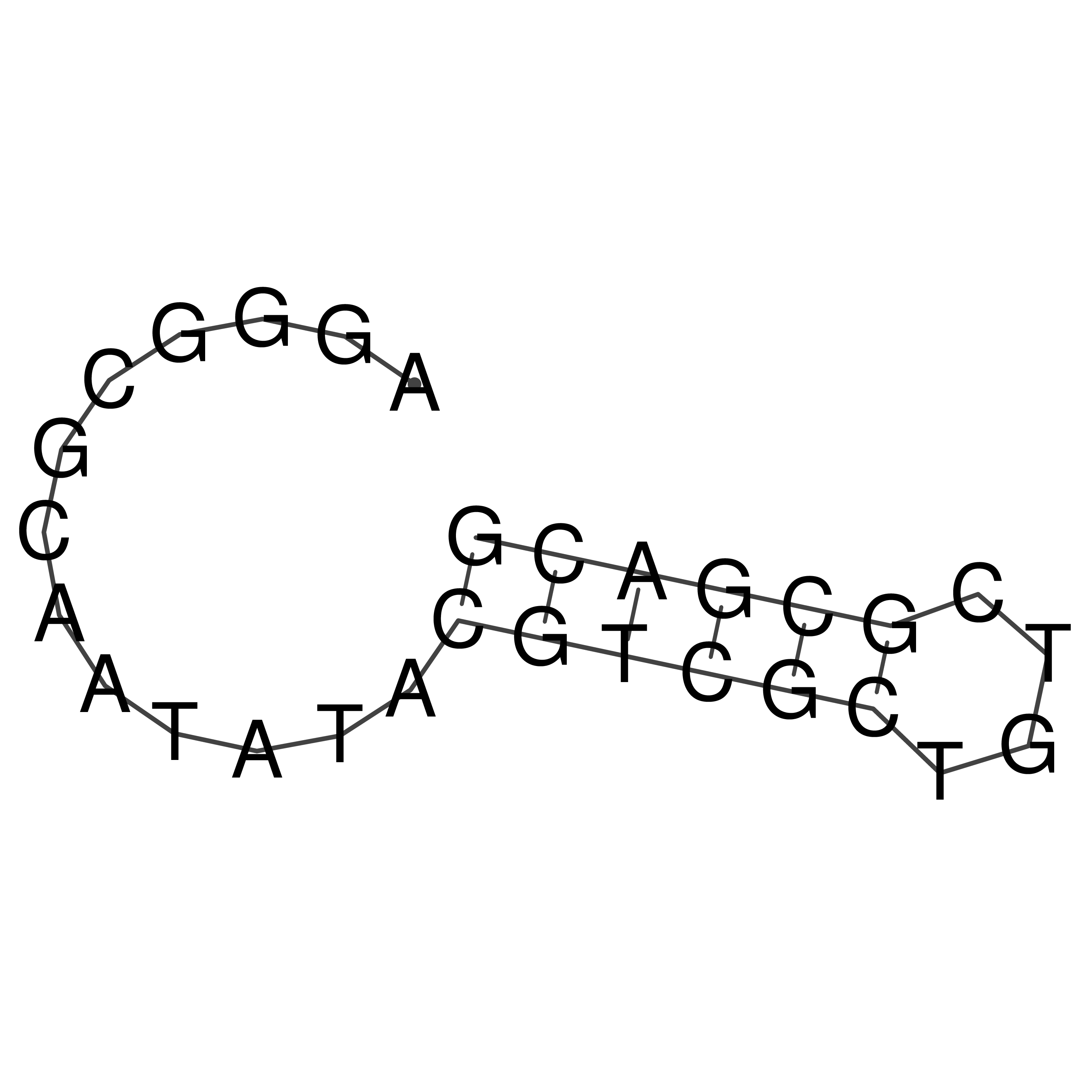
Relaxase
| ID | 2057 | GenBank | WP_046977193 |
| Name | traA_AMK02_RS24250_pRspN731c |
UniProt ID | A0A192TMB6 |
| Length | 1560 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1560 a.a. Molecular weight: 172207.61 Da Isoelectric Point: 6.9611
>WP_046977193.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAGELMYEELALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENITLVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGAKKVPVLGEDGKPLRVVTPDRPNGKIVYKVWAGDKETMKAWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDEREIAKALHRYVDDPVDFANIRVRLMASDQLVILKPQEIEAETGKVSEPAMFTTREMLRIE
YDMAQSARVLSERRGFGVSERNVKVAIESVESGDPKTPFRLDAEQVDAVRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWEGEGHRVIGAALAGKAAEGLQDSSGIKSRTLASWELAWANGRDTLHRGDVLVIDEAGMVA
SQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARNASRLFARGE
VEKGLDAYAQQGHLVEAGTREETIDRIVVDWTEARKHAIGRSISEGRDGRLRGDELLVLAHTNDDVRKLN
EALREVMAGDNALGESRSFRTERGARKFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVVSTGDKR
GGPLLSVLLDNGNKVVFGEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRVDL
YAAIEDFEAKPEWGRKAGVDHAAGVTGELVETGEAKFRPDDEDADDSPYADIRTDGGAVHRLWGVSLPKA
LEEASIMEGDTVTLRKDGVERVKVQIAVVDEKTGHKHYEEREVDRNVWTASQVETASARQERIERESHRP
ELFNLLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEESLTRRWAWIAAKREQVE
KLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHSVSGASADETRYLIPPATSFVSSVEED
ARLAQLASPAWTEREVILRPLLEKIYRDPDAALVSLNALASDIGVAARRLADDLAAAPGRLGRLRGSELI
VDGRAAREERNLAVAAVKELLPMARAHATEFRRNAERFDLREQTRRAHMSLSIPALSERAMARLMEIEAV
RRQGGDDAYKSAFALAAEDRSVVQEIKAVSEALTARFGWSAFSAKADAIAERNIVERMPEDLTDERRGKL
TRLFEAVRRFAEEQHLAERRDRSKVVAGASVDLETEKGVGKENVPIPPMLAAVTEFKVPVDDEARSRALS
APLYRQQRVALANAATTIWRDPAEVVGKIEELLQKGFAADRIGAAVANDPAAYAALRGSDRLVDRMLASG
RERKEAVAAVPEAAARLRALGAAYANALNSERQAITEERRRMGVAIPALSKAAEEALARLTAEARKDSRG
LRVSAVPLDPGIGREFAAVSRALDERFGRNAIVRGEKDLVSRLPPAQRRAFEAMQERLKVLQQTVRLQSS
EQITAERRQRAASRARGINL
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAGELMYEELALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENITLVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGAKKVPVLGEDGKPLRVVTPDRPNGKIVYKVWAGDKETMKAWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDEREIAKALHRYVDDPVDFANIRVRLMASDQLVILKPQEIEAETGKVSEPAMFTTREMLRIE
YDMAQSARVLSERRGFGVSERNVKVAIESVESGDPKTPFRLDAEQVDAVRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWEGEGHRVIGAALAGKAAEGLQDSSGIKSRTLASWELAWANGRDTLHRGDVLVIDEAGMVA
SQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARNASRLFARGE
VEKGLDAYAQQGHLVEAGTREETIDRIVVDWTEARKHAIGRSISEGRDGRLRGDELLVLAHTNDDVRKLN
EALREVMAGDNALGESRSFRTERGARKFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVVSTGDKR
GGPLLSVLLDNGNKVVFGEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRVDL
YAAIEDFEAKPEWGRKAGVDHAAGVTGELVETGEAKFRPDDEDADDSPYADIRTDGGAVHRLWGVSLPKA
LEEASIMEGDTVTLRKDGVERVKVQIAVVDEKTGHKHYEEREVDRNVWTASQVETASARQERIERESHRP
ELFNLLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEESLTRRWAWIAAKREQVE
KLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHSVSGASADETRYLIPPATSFVSSVEED
ARLAQLASPAWTEREVILRPLLEKIYRDPDAALVSLNALASDIGVAARRLADDLAAAPGRLGRLRGSELI
VDGRAAREERNLAVAAVKELLPMARAHATEFRRNAERFDLREQTRRAHMSLSIPALSERAMARLMEIEAV
RRQGGDDAYKSAFALAAEDRSVVQEIKAVSEALTARFGWSAFSAKADAIAERNIVERMPEDLTDERRGKL
TRLFEAVRRFAEEQHLAERRDRSKVVAGASVDLETEKGVGKENVPIPPMLAAVTEFKVPVDDEARSRALS
APLYRQQRVALANAATTIWRDPAEVVGKIEELLQKGFAADRIGAAVANDPAAYAALRGSDRLVDRMLASG
RERKEAVAAVPEAAARLRALGAAYANALNSERQAITEERRRMGVAIPALSKAAEEALARLTAEARKDSRG
LRVSAVPLDPGIGREFAAVSRALDERFGRNAIVRGEKDLVSRLPPAQRRAFEAMQERLKVLQQTVRLQSS
EQITAERRQRAASRARGINL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1713 | GenBank | WP_046977192 |
| Name | traG_AMK02_RS24235_pRspN731c |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70708.95 Da Isoelectric Point: 9.2487
>WP_046977192.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MGLRGKPHPSLLLVLAPIAVTTITIYIVGWRWPGLAVGMSGRMEYWFLRAAPVLPLMFGPLAGLLTVWAL
PLHRRRPVAMASLLCFLGIAAYYALREFGRLAPTVQARVVPWDRALSYLDMVAVIAAVAGFMAVAVSARI
SVVVPDEIKRARRGIFGDADWLPMAAAGKLFPPDGEIVVGERYRVDKEIIHALPFDVNDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALIYTGPMICLDPSTEVAPMVVEHRTNALGREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSGSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNISASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNLSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAK
VNRFVKPVL
MGLRGKPHPSLLLVLAPIAVTTITIYIVGWRWPGLAVGMSGRMEYWFLRAAPVLPLMFGPLAGLLTVWAL
PLHRRRPVAMASLLCFLGIAAYYALREFGRLAPTVQARVVPWDRALSYLDMVAVIAAVAGFMAVAVSARI
SVVVPDEIKRARRGIFGDADWLPMAAAGKLFPPDGEIVVGERYRVDKEIIHALPFDVNDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALIYTGPMICLDPSTEVAPMVVEHRTNALGREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSGSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNISASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNLSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAK
VNRFVKPVL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 190802..200381
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AMK02_RS32280 | 187290..187516 | - | 227 | Protein_180 | chaperonin GroEL | - |
| AMK02_RS24110 (AMK02_PC00164) | 187810..188730 | + | 921 | WP_016737442 | zincin-like metallopeptidase domain-containing protein | - |
| AMK02_RS24115 | 188841..189023 | - | 183 | WP_016737443 | hypothetical protein | - |
| AMK02_RS24120 (AMK02_PC00165) | 189792..190163 | - | 372 | WP_007636193 | hypothetical protein | - |
| AMK02_RS24125 (AMK02_PC00166) | 190262..190798 | + | 537 | WP_016737444 | hypothetical protein | - |
| AMK02_RS24130 (AMK02_PC00167) | 190802..191470 | + | 669 | WP_016737445 | transglycosylase SLT domain-containing protein | virB1 |
| AMK02_RS24135 (AMK02_PC00168) | 191467..191766 | + | 300 | WP_007636189 | TrbC/VirB2 family protein | virB2 |
| AMK02_RS24140 (AMK02_PC00169) | 191772..192110 | + | 339 | WP_011053369 | type IV secretion system protein VirB3 | virB3 |
| AMK02_RS24145 (AMK02_PC00170) | 192103..194469 | + | 2367 | WP_046977188 | VirB4 family type IV secretion system protein | virb4 |
| AMK02_RS24150 (AMK02_PC00171) | 194466..195167 | + | 702 | WP_016737448 | P-type DNA transfer protein VirB5 | virB5 |
| AMK02_RS24155 (AMK02_PC00172) | 195164..195379 | + | 216 | WP_016737449 | EexN family lipoprotein | - |
| AMK02_RS24160 (AMK02_PC00173) | 195382..196314 | + | 933 | WP_046977189 | type IV secretion system protein | virB6 |
| AMK02_RS32285 | 196356..196637 | + | 282 | WP_016737561 | hypothetical protein | - |
| AMK02_RS24165 (AMK02_PC00174) | 196639..197310 | + | 672 | WP_046977190 | virB8 family protein | virB8 |
| AMK02_RS24170 (AMK02_PC00175) | 197307..198164 | + | 858 | WP_016737451 | P-type conjugative transfer protein VirB9 | virB9 |
| AMK02_RS24175 (AMK02_PC00176) | 198173..199345 | + | 1173 | WP_016737452 | type IV secretion system protein VirB10 | virB10 |
| AMK02_RS24180 (AMK02_PC00177) | 199353..200381 | + | 1029 | WP_046977191 | P-type DNA transfer ATPase VirB11 | virB11 |
| AMK02_RS24185 (AMK02_PC00178) | 200425..201003 | - | 579 | WP_016737454 | PIN domain-containing protein | - |
| AMK02_RS24190 (AMK02_PC00179) | 200993..201439 | - | 447 | WP_029532409 | hypothetical protein | - |
| AMK02_RS24195 (AMK02_PC00181) | 201949..203139 | - | 1191 | WP_016737456 | DUF1173 domain-containing protein | - |
| AMK02_RS31415 | 203450..203709 | + | 260 | Protein_200 | toprim domain-containing protein | - |
| AMK02_RS24200 (AMK02_PC00182) | 204032..204972 | + | 941 | Protein_201 | DUF2493 domain-containing protein | - |
| AMK02_RS24205 (AMK02_PC00184) | 205057..205323 | + | 267 | WP_004676093 | hypothetical protein | - |
Host bacterium
| ID | 3228 | GenBank | NZ_CP013604 |
| Plasmid name | pRspN731c | Incompatibility group | - |
| Plasmid size | 433219 bp | Coordinate of oriT [Strand] | 211511..211539 [+] |
| Host baterium | Rhizobium sp. N731 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | hsiC1/vipB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nifX, nifN, nifE, nifK, nifD, nifH, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, nifT, a9174_31165, nodD, nodJ, nodI, nodS, nodC, nodB, nodZ, fixG, fixO, fixN, mLTONO_5203, nodA |
| Anti-CRISPR | - |