Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102782 |
| Name | oriT_pRspN871b |
| Organism | Rhizobium sp. N871 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013592 (215833..215861 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRspN871b
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file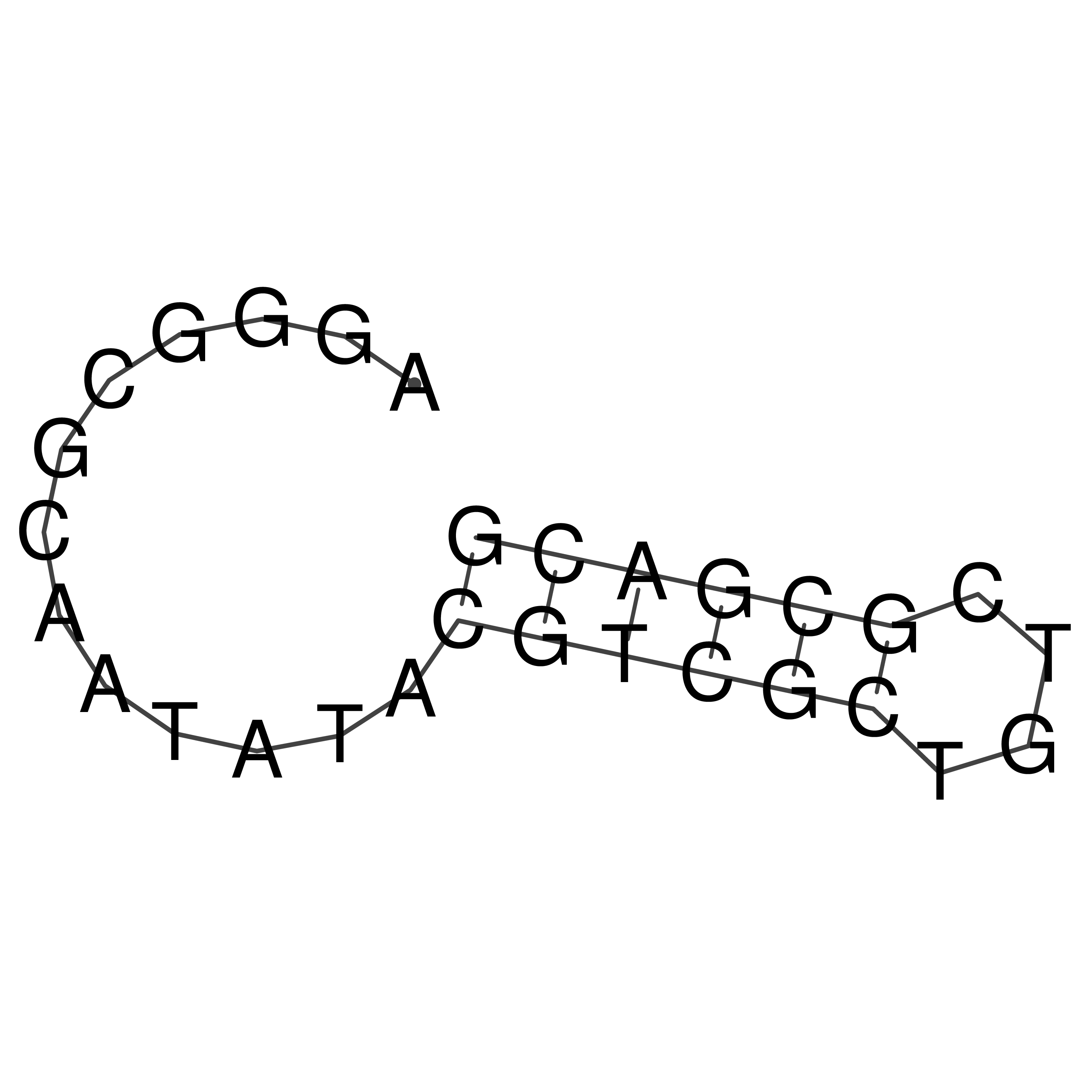
Relaxase
| ID | 2054 | GenBank | WP_064811989 |
| Name | traA_AMK04_RS24035_pRspN871b |
UniProt ID | A0A192M522 |
| Length | 1565 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1565 a.a. Molecular weight: 172913.26 Da Isoelectric Point: 7.2379
>WP_064811989.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAGELMYEELALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENITLVRDFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGAKKVPVLGEGGKPLRVVTPDRPNGKIVYKVWAGDKETLNAWKIAWAETANR
HLALADHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIARALHRYVDDPTDFANIRARLMASDQLVILKPQEIEAETGKVSEPAMFTTREMLRIE
YDMAQSARVLSERRGFGVSERIVTVAIERVESIESGDPKTPFRLDAEQVDAVRHVTGDGDIAAIVGLAGA
GKSTLLAAARLAWEGEGHRVIGAALAGKAAEGLQHSSGIKSRTLASWELAWANGRDTLHRGDVLVIDEAG
MVASQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARNASRLFA
RGEVEKGLDAYARHGHLVEAGSREETIDRIVSDWAAARREAIERSTSEGGDGRLRGDELLVLAHTNDDVR
KLNEALRSVMTQEGALGESRSFRSERGAREFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVVSTG
DKRGDPLLSILLDNGRKLVFSEDSYRHVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDR
VDLYAAKEDFAAKPEWGRKPRVDHAAGVTGELVETGEAKFRPDDEDADDSPYADVRADGGTVHRLWGVSL
PKALEEAGIQEGDTVMLRKDGVERVKVQITVVDEKTGHKHYEEREVDRNVWTASQVETASARQERIERES
HRSELFNPLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEESLTRRWAWIAEKRE
QVAKLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHSVSGASAAETRYLIPPATSFVSSV
EEDARLAQLASPAWTEREVILRPLLQKIYRDPDAALVSLNALASDIGVAPRRLADDLAVAPGRLGRMRGS
ELIVDGRAAREERNVAVAGVKELLPIARAHATEFRRNAERFELREQTRRAHMSLSIPALSERAMARLMEI
EAVRRQGGDDAYKTAFALTAKDRSVVQEIKAVSEALTARFGWSAFSAKADAIAERNIVERMPEDLTDERR
GKLTRLFEAVRRFAEEQHLAERRDRSKIVAGASVDLRTGTEKGVGKENVPIPPMLAAVTEFKVPVDDEAR
SRALASPLYRQQRAALANAATTIWRDPAEVVGKIEELLQKGFAAERIGAAVTNNPAAYGALRGSDRLMDR
MLASGRERKEATRTVPEAAARLRALGAAYANALNSERQAITDERRRMAVAIPALSKAAEEALAHLTVEVS
KDSRKLNVSAASLDPGIGREFAAVSRALDERFGRNALVRGDKDIANVVPPAQRGAFAAMQERLKVLQQTV
RLQSSEQIIVERRQQTANRSRGINL
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAGELMYEELALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENITLVRDFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGAKKVPVLGEGGKPLRVVTPDRPNGKIVYKVWAGDKETLNAWKIAWAETANR
HLALADHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIARALHRYVDDPTDFANIRARLMASDQLVILKPQEIEAETGKVSEPAMFTTREMLRIE
YDMAQSARVLSERRGFGVSERIVTVAIERVESIESGDPKTPFRLDAEQVDAVRHVTGDGDIAAIVGLAGA
GKSTLLAAARLAWEGEGHRVIGAALAGKAAEGLQHSSGIKSRTLASWELAWANGRDTLHRGDVLVIDEAG
MVASQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARNASRLFA
RGEVEKGLDAYARHGHLVEAGSREETIDRIVSDWAAARREAIERSTSEGGDGRLRGDELLVLAHTNDDVR
KLNEALRSVMTQEGALGESRSFRSERGAREFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVVSTG
DKRGDPLLSILLDNGRKLVFSEDSYRHVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDR
VDLYAAKEDFAAKPEWGRKPRVDHAAGVTGELVETGEAKFRPDDEDADDSPYADVRADGGTVHRLWGVSL
PKALEEAGIQEGDTVMLRKDGVERVKVQITVVDEKTGHKHYEEREVDRNVWTASQVETASARQERIERES
HRSELFNPLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEESLTRRWAWIAEKRE
QVAKLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHSVSGASAAETRYLIPPATSFVSSV
EEDARLAQLASPAWTEREVILRPLLQKIYRDPDAALVSLNALASDIGVAPRRLADDLAVAPGRLGRMRGS
ELIVDGRAAREERNVAVAGVKELLPIARAHATEFRRNAERFELREQTRRAHMSLSIPALSERAMARLMEI
EAVRRQGGDDAYKTAFALTAKDRSVVQEIKAVSEALTARFGWSAFSAKADAIAERNIVERMPEDLTDERR
GKLTRLFEAVRRFAEEQHLAERRDRSKIVAGASVDLRTGTEKGVGKENVPIPPMLAAVTEFKVPVDDEAR
SRALASPLYRQQRAALANAATTIWRDPAEVVGKIEELLQKGFAAERIGAAVTNNPAAYGALRGSDRLMDR
MLASGRERKEATRTVPEAAARLRALGAAYANALNSERQAITDERRRMAVAIPALSKAAEEALAHLTVEVS
KDSRKLNVSAASLDPGIGREFAAVSRALDERFGRNALVRGDKDIANVVPPAQRGAFAAMQERLKVLQQTV
RLQSSEQIIVERRQQTANRSRGINL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1710 | GenBank | WP_064811988 |
| Name | traG_AMK04_RS24020_pRspN871b |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70745.97 Da Isoelectric Point: 9.5675
>WP_064811988.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MGLRGKPHPSLLLVLVPIAVTTITIYIVGWRWPGLAAGMSGKIEHWFLRAAPVPPLLFGPLAGLLTVWAL
PLHRRRPVAMASLLYFLGVAAFYALREFGRLAPAVQAEVITWDRALSYLDMVAVIAAVAGFMAVAMSARI
SVVVPDEIKRARRGIFGDADWLPMRAAGKLFPPEGEIVVGERYRVDKEIVHALPFDANDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYSGPLICLDPSTEVAPMVAEHRARALKREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVTFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAK
VNRFVKPVL
MGLRGKPHPSLLLVLVPIAVTTITIYIVGWRWPGLAAGMSGKIEHWFLRAAPVPPLLFGPLAGLLTVWAL
PLHRRRPVAMASLLYFLGVAAFYALREFGRLAPAVQAEVITWDRALSYLDMVAVIAAVAGFMAVAMSARI
SVVVPDEIKRARRGIFGDADWLPMRAAGKLFPPEGEIVVGERYRVDKEIVHALPFDANDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYSGPLICLDPSTEVAPMVAEHRARALKREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVTFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAK
VNRFVKPVL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 196072..205660
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AMK04_RS33680 | 192556..192782 | - | 227 | Protein_184 | chaperonin GroEL | - |
| AMK04_RS23895 (AMK04_PB00172) | 193077..193997 | + | 921 | WP_064811974 | zincin-like metallopeptidase domain-containing protein | - |
| AMK04_RS23900 | 194108..194290 | - | 183 | WP_016737443 | hypothetical protein | - |
| AMK04_RS23905 (AMK04_PB00173) | 195062..195433 | - | 372 | WP_042120136 | hypothetical protein | - |
| AMK04_RS23910 (AMK04_PB00174) | 195532..196068 | + | 537 | WP_064811975 | hypothetical protein | - |
| AMK04_RS23915 (AMK04_PB00175) | 196072..196731 | + | 660 | WP_064811976 | lytic transglycosylase domain-containing protein | virB1 |
| AMK04_RS23920 (AMK04_PB00176) | 196728..197027 | + | 300 | WP_009997887 | TrbC/VirB2 family protein | virB2 |
| AMK04_RS23925 (AMK04_PB00177) | 197033..197371 | + | 339 | WP_009997888 | type IV secretion system protein VirB3 | virB3 |
| AMK04_RS23930 (AMK04_PB00178) | 197364..199730 | + | 2367 | WP_064811977 | VirB4 family type IV secretion system protein | virb4 |
| AMK04_RS23935 (AMK04_PB00179) | 199727..200428 | + | 702 | WP_064811978 | P-type DNA transfer protein VirB5 | virB5 |
| AMK04_RS23940 (AMK04_PB00180) | 200425..200658 | + | 234 | WP_010007459 | EexN family lipoprotein | - |
| AMK04_RS23945 (AMK04_PB00181) | 200662..201594 | + | 933 | WP_040111793 | type IV secretion system protein | virB6 |
| AMK04_RS33685 (AMK04_PB00182) | 201634..201915 | + | 282 | WP_080771758 | hypothetical protein | - |
| AMK04_RS23950 (AMK04_PB00183) | 201917..202588 | + | 672 | WP_064811979 | virB8 family protein | virB8 |
| AMK04_RS23955 (AMK04_PB00184) | 202585..203442 | + | 858 | WP_064811980 | P-type conjugative transfer protein VirB9 | virB9 |
| AMK04_RS23960 (AMK04_PB00185) | 203451..204623 | + | 1173 | WP_064811981 | type IV secretion system protein VirB10 | virB10 |
| AMK04_RS23965 (AMK04_PB00186) | 204632..205660 | + | 1029 | WP_010008207 | P-type DNA transfer ATPase VirB11 | virB11 |
| AMK04_RS23970 (AMK04_PB00187) | 205704..206282 | - | 579 | WP_064811982 | PIN domain-containing protein | - |
| AMK04_RS23975 (AMK04_PB00188) | 206272..206718 | - | 447 | WP_225881772 | DNA-binding protein | - |
| AMK04_RS23980 (AMK04_PB00190) | 207230..208420 | - | 1191 | WP_064811984 | DUF1173 domain-containing protein | - |
| AMK04_RS33690 | 208731..208990 | + | 260 | Protein_204 | toprim domain-containing protein | - |
| AMK04_RS23990 (AMK04_PB00191) | 209376..209642 | + | 267 | WP_004676093 | hypothetical protein | - |
| AMK04_RS23995 (AMK04_PB00192) | 209639..210037 | + | 399 | WP_064811986 | type II toxin-antitoxin system VapC family toxin | - |
Host bacterium
| ID | 3225 | GenBank | NZ_CP013592 |
| Plasmid name | pRspN871b | Incompatibility group | - |
| Plasmid size | 433056 bp | Coordinate of oriT [Strand] | 215833..215861 [+] |
| Host baterium | Rhizobium sp. N871 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | hsiC1/vipB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nifX, nifN, nifE, nifK, nifD, nifH, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, nifT, nodD, nodJ, nodI, nodS, nodC, fixO, fixN, mLTONO_5203, nodA |
| Anti-CRISPR | - |