Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102781 |
| Name | oriT_pRphaR620d |
| Organism | Rhizobium phaseoli strain R620 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013546 (962481..962509 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRphaR620d
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file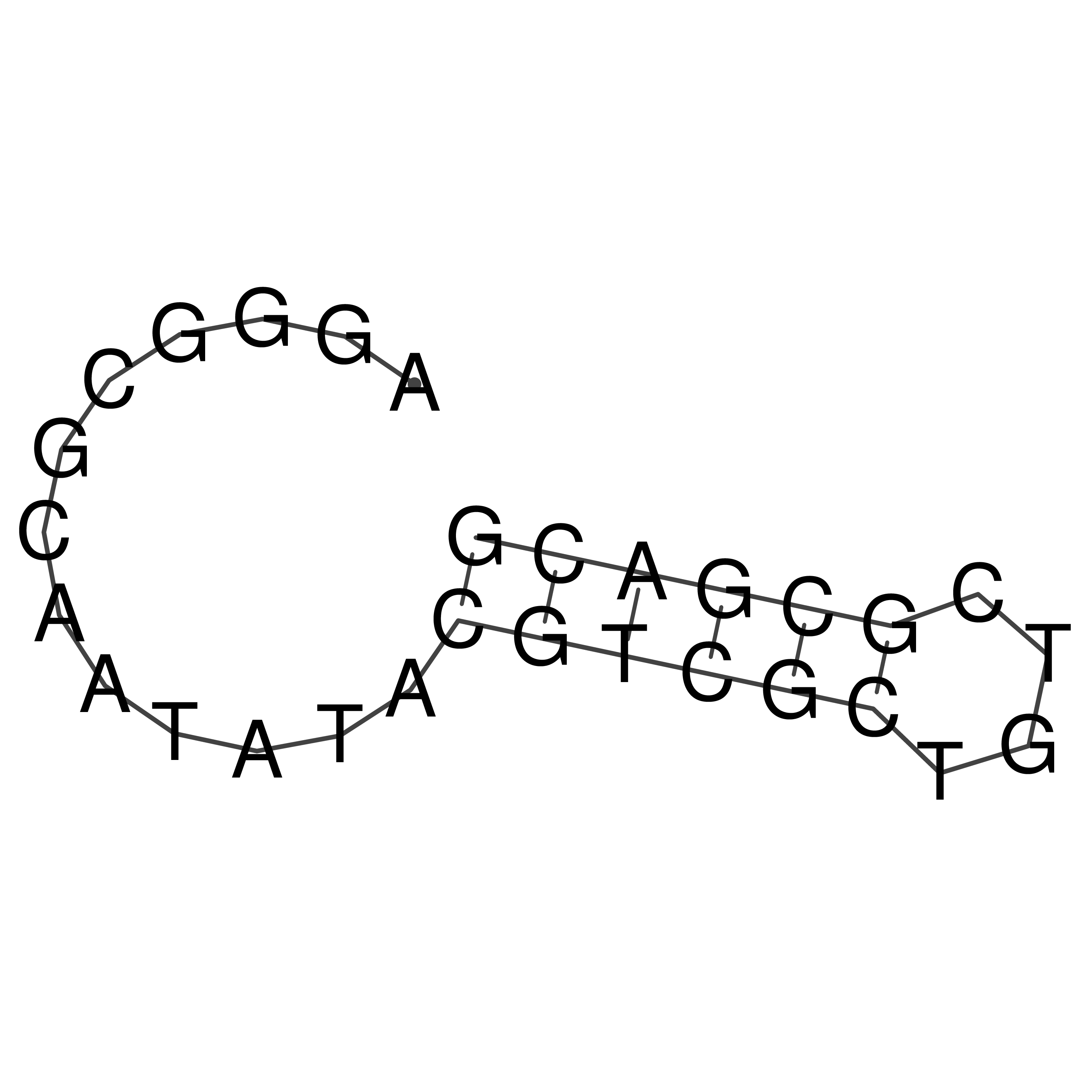
Relaxase
| ID | 2053 | GenBank | WP_064826469 |
| Name | traA_AMC86_RS31005_pRphaR620d |
UniProt ID | _ |
| Length | 1536 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1536 a.a. Molecular weight: 170773.15 Da Isoelectric Point: 7.2193
>WP_064826469.1 Ti-type conjugative transfer relaxase TraA [Rhizobium phaseoli]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGAASELMHEELALPDQIPAWLKMAMDG
RSIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEHSKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGVAQKHLGPEKAALARKGRELFFAPADLARRQGMADRLLAEPELLLKQ
LGNERSTFDEKDIAKALHRCVDDPVEFANIRARLMASDELVMLKPQEMDADTGKVSEPAVFTTRDMLRIE
YHMAESARRLSNRSGYAVSRKHVAAAIERIESEDPKNLFHLDAEQVDAIRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAEIEVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VEKGLDAYAQQGHLVEAGTREEAINRIVADWTEARKQAIDRSTSEGRDGHLRGDELLVLAHTNADVHKLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPLMSVLLDNGRNVVFSEDSYRNVDHGYASTIHKSQGATVDRTFVLATGMMDRHLTYVSMTRHRDRADL
YAVKEDFQVRSERGRRLRADHAAGVTGELVEAGEAKFRPDDEDADDSPYADIRTDDGTVHRLWGVSLPKA
LEEAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYEEREVGRNVWTAKQIETAEARQERIERESHRP
DVFSRLVERLSRSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGLAIERERRVAYNEERSETMIGAISSEERYLIPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASDTGTEPRKLADDLAVAPDLLGRLYGSDLIVDGRAARDERNVAMA
TVKELLPLARAHATEFRRNATRFERREQTRRNYMVLSIPALSERAVARLTEIEAVRSRGGDDAYKTASAL
AAQDRTVVQEIKAVSEAITARFGSSAFTAKADAIAQRNMAERMLEELTSVRGEKLIRLFEAVRRFAEEQH
LAERQDRSKIVAGASADPQNQNVPMLAAVTEFKTPIDEEARSRVLAGPLYRQQRAALAAVASTIWRDPAG
AVDKIEELLVKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVPEAAARLRSLGSAY
VNVLDAERQAIAEERRRMAVAIPGLSKSAEEVLMRLTTEAKNNGRTRNAAAASLDPSIHREFEAVSRALD
ERFGHRAIIRGEKDLFNRMPLAQRRAFEVMWERFKVLQQTVRWESSEKIVSELRNRTINRGRGIDL
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGAASELMHEELALPDQIPAWLKMAMDG
RSIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEHSKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGVAQKHLGPEKAALARKGRELFFAPADLARRQGMADRLLAEPELLLKQ
LGNERSTFDEKDIAKALHRCVDDPVEFANIRARLMASDELVMLKPQEMDADTGKVSEPAVFTTRDMLRIE
YHMAESARRLSNRSGYAVSRKHVAAAIERIESEDPKNLFHLDAEQVDAIRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAEIEVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VEKGLDAYAQQGHLVEAGTREEAINRIVADWTEARKQAIDRSTSEGRDGHLRGDELLVLAHTNADVHKLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPLMSVLLDNGRNVVFSEDSYRNVDHGYASTIHKSQGATVDRTFVLATGMMDRHLTYVSMTRHRDRADL
YAVKEDFQVRSERGRRLRADHAAGVTGELVEAGEAKFRPDDEDADDSPYADIRTDDGTVHRLWGVSLPKA
LEEAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYEEREVGRNVWTAKQIETAEARQERIERESHRP
DVFSRLVERLSRSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGLAIERERRVAYNEERSETMIGAISSEERYLIPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASDTGTEPRKLADDLAVAPDLLGRLYGSDLIVDGRAARDERNVAMA
TVKELLPLARAHATEFRRNATRFERREQTRRNYMVLSIPALSERAVARLTEIEAVRSRGGDDAYKTASAL
AAQDRTVVQEIKAVSEAITARFGSSAFTAKADAIAQRNMAERMLEELTSVRGEKLIRLFEAVRRFAEEQH
LAERQDRSKIVAGASADPQNQNVPMLAAVTEFKTPIDEEARSRVLAGPLYRQQRAALAAVASTIWRDPAG
AVDKIEELLVKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVPEAAARLRSLGSAY
VNVLDAERQAIAEERRRMAVAIPGLSKSAEEVLMRLTTEAKNNGRTRNAAAASLDPSIHREFEAVSRALD
ERFGHRAIIRGEKDLFNRMPLAQRRAFEVMWERFKVLQQTVRWESSEKIVSELRNRTINRGRGIDL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1709 | GenBank | WP_037146751 |
| Name | traG_AMC86_RS31020_pRphaR620d |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70465.33 Da Isoelectric Point: 9.2555
>WP_037146751.1 Ti-type conjugative transfer system protein TraG [Rhizobium phaseoli]
MIARGKPHPSLLFVFAPVVVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVFASYAMREFARLGPSVQTGVITWDRALSYLDMVAVLGAVAGFLAVAVSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDANDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHALGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQVIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
MIARGKPHPSLLFVFAPVVVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVFASYAMREFARLGPSVQTGVITWDRALSYLDMVAVLGAVAGFLAVAVSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDANDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHALGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQVIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 973784..983339
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AMC86_RS31050 | 969355..969537 | + | 183 | WP_037146733 | hypothetical protein | - |
| AMC86_RS31055 (AMC86_PD00894) | 969670..970710 | - | 1041 | WP_037146729 | toprim domain-containing protein | - |
| AMC86_RS31060 (AMC86_PD00895) | 971073..972263 | + | 1191 | WP_037146725 | DUF1173 domain-containing protein | - |
| AMC86_RS31065 (AMC86_PD00896) | 972706..973299 | + | 594 | WP_037146723 | DUF433 domain-containing protein | - |
| AMC86_RS31070 (AMC86_PD00897) | 973415..973765 | + | 351 | WP_237353383 | type II toxin-antitoxin system VapC family toxin | - |
| AMC86_RS31075 (AMC86_PD00898) | 973784..974779 | - | 996 | WP_037146721 | P-type DNA transfer ATPase VirB11 | virB11 |
| AMC86_RS31080 (AMC86_PD00899) | 974788..975963 | - | 1176 | WP_016735275 | type IV secretion system protein VirB10 | virB10 |
| AMC86_RS31085 (AMC86_PD00900) | 975972..976826 | - | 855 | WP_037146719 | P-type conjugative transfer protein VirB9 | virB9 |
| AMC86_RS31090 (AMC86_PD00901) | 976823..977494 | - | 672 | WP_037146717 | virB8 family protein | virB8 |
| AMC86_RS34075 (AMC86_PD00902) | 977488..977775 | - | 288 | WP_080712055 | hypothetical protein | - |
| AMC86_RS31095 (AMC86_PD00903) | 977815..978747 | - | 933 | WP_037146716 | type IV secretion system protein | virB6 |
| AMC86_RS31100 (AMC86_PD00904) | 978750..978983 | - | 234 | WP_016735278 | EexN family lipoprotein | - |
| AMC86_RS31105 (AMC86_PD00905) | 978980..979681 | - | 702 | WP_016735279 | P-type DNA transfer protein VirB5 | virB5 |
| AMC86_RS31110 (AMC86_PD00906) | 979678..982044 | - | 2367 | WP_064826470 | VirB4 family type IV secretion system protein | virb4 |
| AMC86_RS31115 (AMC86_PD00907) | 982037..982375 | - | 339 | WP_016735282 | type IV secretion system protein VirB3 | virB3 |
| AMC86_RS31120 (AMC86_PD00908) | 982381..982680 | - | 300 | WP_016735283 | TrbC/VirB2 family protein | virB2 |
| AMC86_RS31125 (AMC86_PD00909) | 982677..983339 | - | 663 | WP_037147233 | transglycosylase SLT domain-containing protein | virB1 |
| AMC86_RS31130 (AMC86_PD00910) | 983343..983879 | - | 537 | WP_016735285 | hypothetical protein | - |
| AMC86_RS31135 (AMC86_PD00911) | 983978..984349 | + | 372 | WP_016735286 | hypothetical protein | - |
| AMC86_RS31140 (AMC86_PD00912) | 984702..985892 | - | 1191 | WP_037147230 | hypothetical protein | - |
| AMC86_RS31145 (AMC86_PD00913) | 985968..986624 | - | 657 | WP_037147226 | hypothetical protein | - |
Host bacterium
| ID | 3224 | GenBank | NZ_CP013546 |
| Plasmid name | pRphaR620d | Incompatibility group | - |
| Plasmid size | 1080727 bp | Coordinate of oriT [Strand] | 962481..962509 [-] |
| Host baterium | Rhizobium phaseoli strain R620 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | hupE2 |
| Degradation gene | dxnH, LinG |
| Symbiosis gene | noeL, fixB, fixA |
| Anti-CRISPR | AcrIIA8 |