Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102778 |
| Name | oriT_pRphaN161d |
| Organism | Rhizobium phaseoli strain N161 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013589 (936266..936294 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRphaN161d
AGGGCGCAATATACGTCGCTGCCGCGACG
AGGGCGCAATATACGTCGCTGCCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file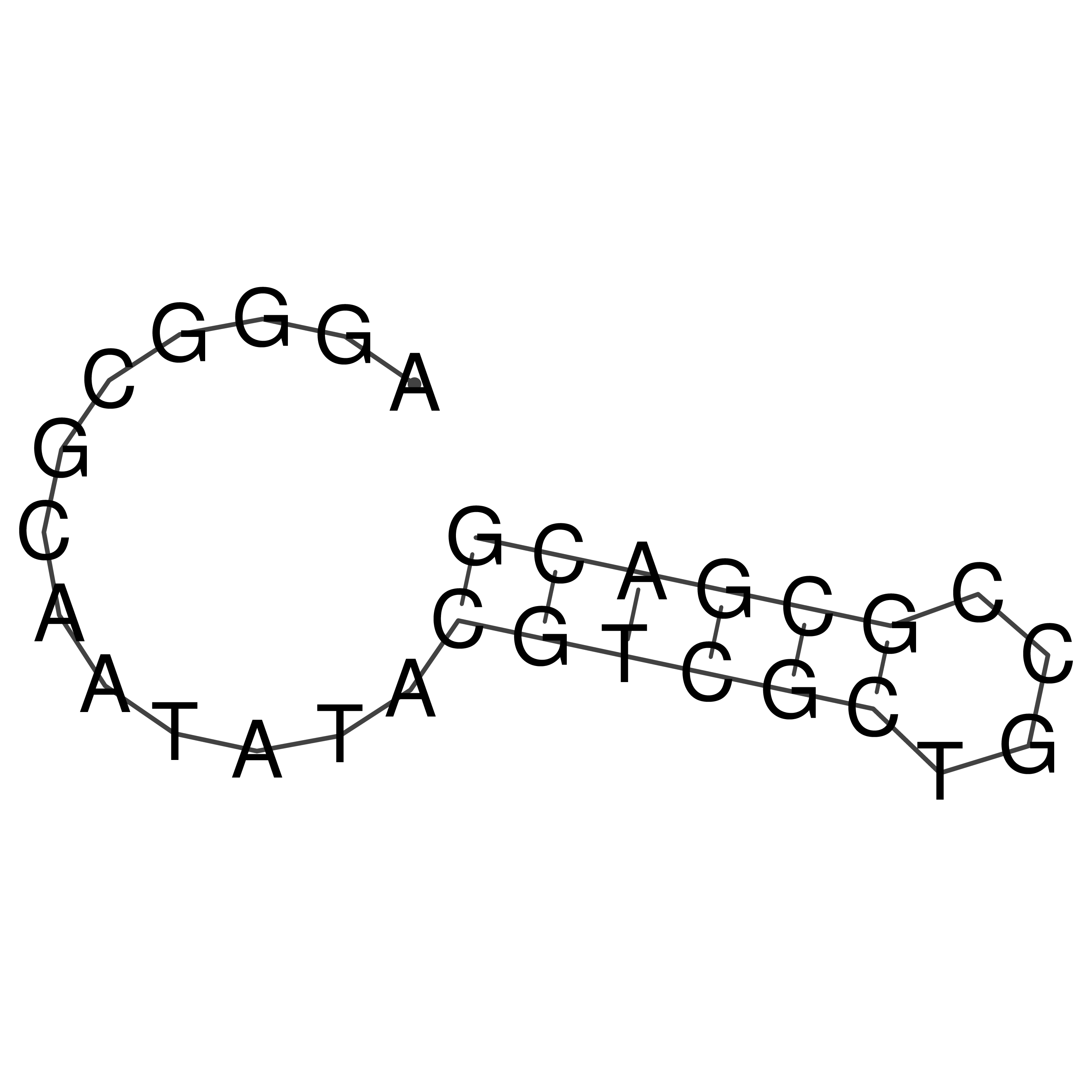
Relaxase
| ID | 2050 | GenBank | WP_064837782 |
| Name | traA_AMC78_RS31415_pRphaN161d |
UniProt ID | _ |
| Length | 1545 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1545 a.a. Molecular weight: 171311.65 Da Isoelectric Point: 6.7554
>WP_064837782.1 Ti-type conjugative transfer relaxase TraA [Rhizobium phaseoli]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGISFSYRGGASELMHEELALPDQIPAWLKMAMDG
RSIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVSGNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEHGKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELFFAPADLARRQEMADRLLAEPELLLKQ
LGNESSTFDEKDIAKALHRYVDDPVEFANIRARLMASDELVMLKPQEMDADTGKVSEPALFTTRDMLRIE
YYMADSARRLSNRSGYAVSRKHVAAAIERIESDDPKNLFRLDAEQGDAIRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVMVIDEAGMVS
SQQMARVLKIAEEAEIKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VERGLDAYAQQGHLVEAGTREEAINRIVVDWTEARKQAIDRSTSEGRDGHLRGDELLVLAHTNADVKRLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPMLSVLLDNGRNVVFSEDSYGNVDHGYAATIHKSQGATVDRTFVLATGIMDRHLTYVSMTRHRDRADL
YAAKEDFQARSEWGRRLRADHAAGVTGELVETGEAKFRPDDEDADDSPYADIRTDDGTVHRLWGVSLPKA
LEEAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYEEREVDRNVWTAKQIETAEARQERIEREGHRP
DVFGRLVERLARSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGLAIERERRVAYNEERSETMIGAISSEERYLMPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASDTGTEPRKLADDLAVAPDLLGRLHGSDLIVDGRAAREERDVAMA
ALAELLPLARGHATEFRRNAERFELREQTRRASMSLSVPALSERAMTRLLEIEAVRTQGGDDAYKTAFAL
AAEDRSVVQEVKAVSEALTARFGWSAFTAKADAMAERNMVERMPEDLSAGRREELTRLFEAVKRFDGEQH
LAERQDRWRIVAGAAAVPGTEQGKEQRKENVTIPPMLAAVTEFKTPVDEEARSRVLSAPLYRQQRVALAN
AARTIWRDPAGAVDKIEELLVKGFAADRIAPAVTNDPAAYGALRGSGRLMDRMLASGRERKEAVQAVPEA
AARLRSLGSAYVNVLDAERQAIAEERRRMAVAIPGLSKAAEGVLMRLTTEAKNNGRTRNAATGSLDPSIH
REFDAVSRALDERFGRSARGEKDLLNRMPPAQRAAFAAMQEKLKVLQQTVRWESSEKIVSELRNRTVNRG
RDIDL
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGISFSYRGGASELMHEELALPDQIPAWLKMAMDG
RSIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVSGNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEHGKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELFFAPADLARRQEMADRLLAEPELLLKQ
LGNESSTFDEKDIAKALHRYVDDPVEFANIRARLMASDELVMLKPQEMDADTGKVSEPALFTTRDMLRIE
YYMADSARRLSNRSGYAVSRKHVAAAIERIESDDPKNLFRLDAEQGDAIRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVMVIDEAGMVS
SQQMARVLKIAEEAEIKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VERGLDAYAQQGHLVEAGTREEAINRIVVDWTEARKQAIDRSTSEGRDGHLRGDELLVLAHTNADVKRLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPMLSVLLDNGRNVVFSEDSYGNVDHGYAATIHKSQGATVDRTFVLATGIMDRHLTYVSMTRHRDRADL
YAAKEDFQARSEWGRRLRADHAAGVTGELVETGEAKFRPDDEDADDSPYADIRTDDGTVHRLWGVSLPKA
LEEAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYEEREVDRNVWTAKQIETAEARQERIEREGHRP
DVFGRLVERLARSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGLAIERERRVAYNEERSETMIGAISSEERYLMPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASDTGTEPRKLADDLAVAPDLLGRLHGSDLIVDGRAAREERDVAMA
ALAELLPLARGHATEFRRNAERFELREQTRRASMSLSVPALSERAMTRLLEIEAVRTQGGDDAYKTAFAL
AAEDRSVVQEVKAVSEALTARFGWSAFTAKADAMAERNMVERMPEDLSAGRREELTRLFEAVKRFDGEQH
LAERQDRWRIVAGAAAVPGTEQGKEQRKENVTIPPMLAAVTEFKTPVDEEARSRVLSAPLYRQQRVALAN
AARTIWRDPAGAVDKIEELLVKGFAADRIAPAVTNDPAAYGALRGSGRLMDRMLASGRERKEAVQAVPEA
AARLRSLGSAYVNVLDAERQAIAEERRRMAVAIPGLSKAAEGVLMRLTTEAKNNGRTRNAATGSLDPSIH
REFDAVSRALDERFGRSARGEKDLLNRMPPAQRAAFAAMQEKLKVLQQTVRWESSEKIVSELRNRTVNRG
RDIDL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1705 | GenBank | WP_064837787 |
| Name | traG_AMC78_RS31430_pRphaN161d |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70461.39 Da Isoelectric Point: 9.3885
>WP_064837787.1 Ti-type conjugative transfer system protein TraG [Rhizobium phaseoli]
MIARGKPHPSLLFVFAPVVVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVSASFAMREFARLGPSVQTGVITWDRALSYLDMVAVLGAVAGFLAVALSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDANDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHTLGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GKAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
MIARGKPHPSLLFVFAPVVVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVSASFAMREFARLGPSVQTGVITWDRALSYLDMVAVLGAVAGFLAVALSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDANDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHTLGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GKAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 947555..957110
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AMC78_RS31460 | 943125..943307 | + | 183 | WP_037146733 | hypothetical protein | - |
| AMC78_RS31465 (AMC78_PD00861) | 943440..944480 | - | 1041 | WP_037146729 | toprim domain-containing protein | - |
| AMC78_RS31470 (AMC78_PD00862) | 944843..946033 | + | 1191 | WP_037146725 | DUF1173 domain-containing protein | - |
| AMC78_RS31475 (AMC78_PD00863) | 946476..947069 | + | 594 | WP_037146723 | DUF433 domain-containing protein | - |
| AMC78_RS31480 (AMC78_PD00864) | 947084..947536 | + | 453 | WP_016735273 | PIN domain-containing protein | - |
| AMC78_RS31485 (AMC78_PD00865) | 947555..948550 | - | 996 | WP_064829760 | P-type DNA transfer ATPase VirB11 | virB11 |
| AMC78_RS31490 (AMC78_PD00866) | 948559..949734 | - | 1176 | WP_064837789 | type IV secretion system protein VirB10 | virB10 |
| AMC78_RS31495 (AMC78_PD00867) | 949743..950597 | - | 855 | WP_064837791 | P-type conjugative transfer protein VirB9 | virB9 |
| AMC78_RS31500 (AMC78_PD00868) | 950594..951265 | - | 672 | WP_064837792 | virB8 family protein | virB8 |
| AMC78_RS34755 (AMC78_PD00869) | 951259..951546 | - | 288 | WP_080712055 | hypothetical protein | - |
| AMC78_RS31505 (AMC78_PD00870) | 951586..952518 | - | 933 | WP_064837794 | type IV secretion system protein | virB6 |
| AMC78_RS31510 (AMC78_PD00871) | 952521..952754 | - | 234 | WP_012490331 | EexN family lipoprotein | - |
| AMC78_RS31515 (AMC78_PD00872) | 952751..953452 | - | 702 | WP_064837796 | P-type DNA transfer protein VirB5 | virB5 |
| AMC78_RS31520 (AMC78_PD00873) | 953449..955815 | - | 2367 | WP_064837797 | VirB4 family type IV secretion system protein | virb4 |
| AMC78_RS31525 (AMC78_PD00874) | 955808..956146 | - | 339 | WP_064837799 | type IV secretion system protein VirB3 | virB3 |
| AMC78_RS31530 (AMC78_PD00875) | 956152..956451 | - | 300 | WP_016735283 | TrbC/VirB2 family protein | virB2 |
| AMC78_RS31535 (AMC78_PD00876) | 956448..957110 | - | 663 | WP_064837801 | transglycosylase SLT domain-containing protein | virB1 |
| AMC78_RS31540 (AMC78_PD00877) | 957114..957650 | - | 537 | WP_064820513 | hypothetical protein | - |
| AMC78_RS31545 (AMC78_PD00878) | 957749..958120 | + | 372 | WP_064824316 | hypothetical protein | - |
| AMC78_RS31550 (AMC78_PD00879) | 958473..959663 | - | 1191 | WP_064837803 | hypothetical protein | - |
| AMC78_RS31555 (AMC78_PD00880) | 959737..960393 | - | 657 | WP_064830295 | hypothetical protein | - |
Host bacterium
| ID | 3221 | GenBank | NZ_CP013589 |
| Plasmid name | pRphaN161d | Incompatibility group | - |
| Plasmid size | 1048055 bp | Coordinate of oriT [Strand] | 936266..936294 [-] |
| Host baterium | Rhizobium phaseoli strain N161 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | hupE2 |
| Degradation gene | dxnH, LinG |
| Symbiosis gene | noeL, fixB, fixA |
| Anti-CRISPR | AcrIIA8 |