Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102777 |
| Name | oriT_pRphaN161b |
| Organism | Rhizobium phaseoli strain N161 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013587 (177632..177660 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRphaN161b
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file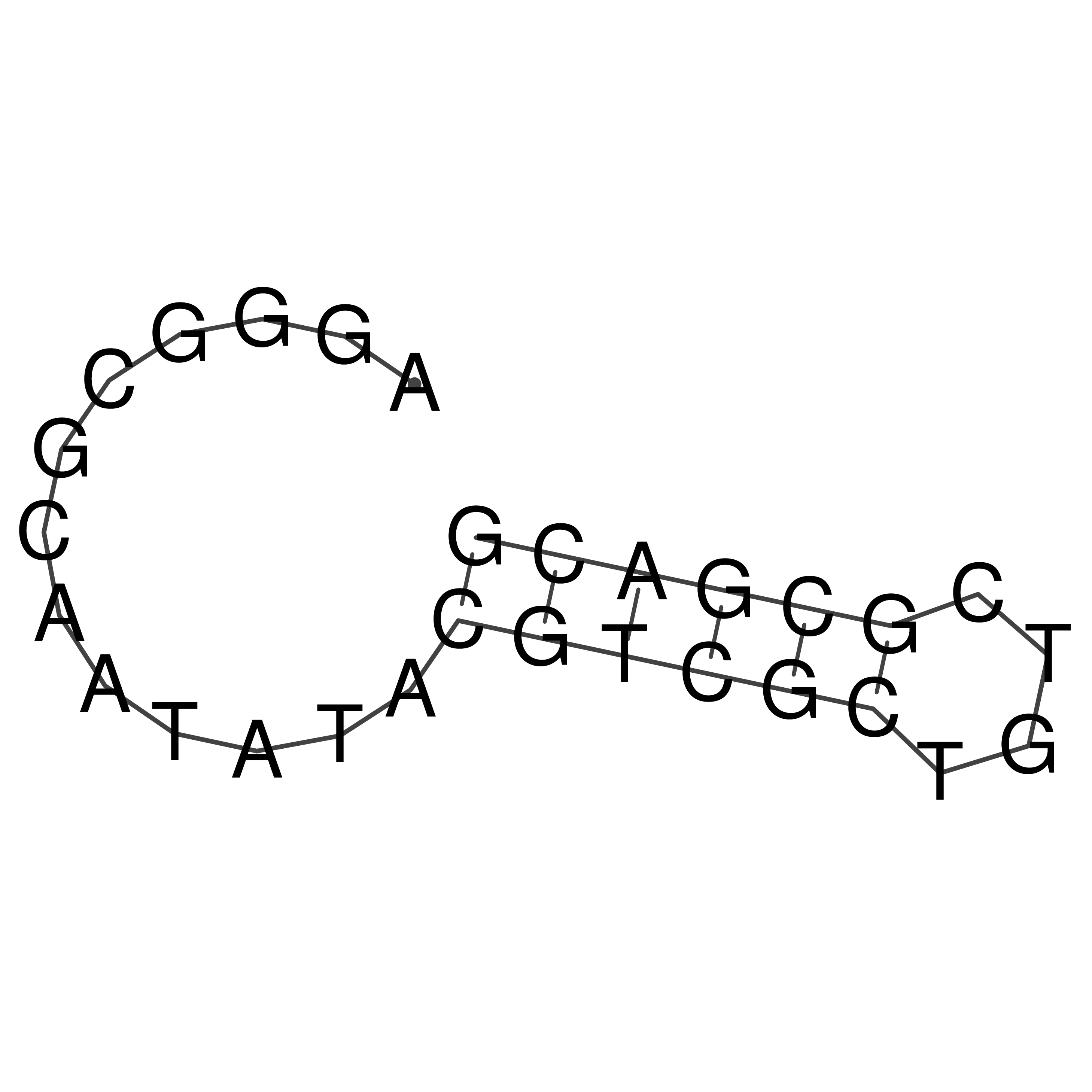
Relaxase
| ID | 2049 | GenBank | WP_029875464 |
| Name | traA_AMC78_RS24305_pRphaN161b |
UniProt ID | A0A2A5KKZ8 |
| Length | 1558 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1558 a.a. Molecular weight: 172030.24 Da Isoelectric Point: 7.1540
>WP_029875464.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAGELMYEELALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENITLVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPATEEGFGAKKVPVLGEGGKPLRVVTPDRPNGKIVYKVWAGDKETMKAWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDERDIARALHRYVDDPTDFANIRARLMASDQLVILKPQEIEAETGKVSEPAVFTTREMLRIE
YDMAQSARVLSERRGFGVSERNVTVAIERVESGDPKNPFRLDAEQVDAVRHVTGDGGIAAIVGLAGAGKS
TLLAAARLAWESEGHRVIGAALAGKAAEGLQDSSGIKSRTLASWELAWGNGRDTLHRGDVLVIDEAGMVA
SQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELVGVRRQREAWARNASRLFARGE
VEKGLDAYARHGHLVEAGSREETIDRIVSDWAAARREAIERSTSEGGDGRLRGDELLVLAHTNDDVRKLN
EALRSVMTQEGALSESRSFRSERGVREFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVVSTGDKR
GDPLLSVLLDNGRKLVFSEDSYRHVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRVDL
YAAKEDFAAKPEWGRKPRVDHATGVTGELVETGEAKFRPDDEDADDSPYADVRADDGTVHRLWGVSLPKA
LEEAGIQEGDTVTLRKDGVERVKVQIAVVDEKTGHKHYEEREVDRNVWTASQVETASARQERIERESHRP
ELFNPLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEQSLTRRWAWIAAKREQVE
KLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHSVSGASAAETRYLIPPATSFVSSVEED
ARLAQLASPAWTEREVILRPLLQKIYRDPDAALVSLNALASDIGVAPRRLADDLAAAPGRLGRLRGSELI
VDGRAAREERNLAVAAVKELLPMARAHATEFRRNAERFELREQTRRAHMSLSIPALSERAMARLMEIEAV
RSQGGDDAYKTAFALAAKDRSVVREIKAVSEALTARFGWSAFSAKADAIAERNIVERMPEDLTDEWRGKL
TRLFDAVRRFADEQHLAERRDRSKVVAGASADLSKEPGTEKIIMPPMFAAVTEFKVPIDDEARSRALASP
VYRQQRAALANAATTIWRDPAEVVGKIEELLQKGFAAERIGAAVTNNPAAYGALRGSDRLMDRMLTSGRE
RKEAVAAVPEAAARLRALGAAHLNALDAERQAITDERRRMAVAIPALSKAAEEALAHLTVEVSKDSRKLS
VSAASLDPGIGREFAAVSRALDERFGRNALVRGDKDIANVVPPAQRGAFAAMQERLKVLQQTVRLQSSEQ
IIVERRQQTANRSRGINL
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAGELMYEELALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENITLVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPATEEGFGAKKVPVLGEGGKPLRVVTPDRPNGKIVYKVWAGDKETMKAWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDERDIARALHRYVDDPTDFANIRARLMASDQLVILKPQEIEAETGKVSEPAVFTTREMLRIE
YDMAQSARVLSERRGFGVSERNVTVAIERVESGDPKNPFRLDAEQVDAVRHVTGDGGIAAIVGLAGAGKS
TLLAAARLAWESEGHRVIGAALAGKAAEGLQDSSGIKSRTLASWELAWGNGRDTLHRGDVLVIDEAGMVA
SQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELVGVRRQREAWARNASRLFARGE
VEKGLDAYARHGHLVEAGSREETIDRIVSDWAAARREAIERSTSEGGDGRLRGDELLVLAHTNDDVRKLN
EALRSVMTQEGALSESRSFRSERGVREFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVVSTGDKR
GDPLLSVLLDNGRKLVFSEDSYRHVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRVDL
YAAKEDFAAKPEWGRKPRVDHATGVTGELVETGEAKFRPDDEDADDSPYADVRADDGTVHRLWGVSLPKA
LEEAGIQEGDTVTLRKDGVERVKVQIAVVDEKTGHKHYEEREVDRNVWTASQVETASARQERIERESHRP
ELFNPLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEQSLTRRWAWIAAKREQVE
KLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHSVSGASAAETRYLIPPATSFVSSVEED
ARLAQLASPAWTEREVILRPLLQKIYRDPDAALVSLNALASDIGVAPRRLADDLAAAPGRLGRLRGSELI
VDGRAAREERNLAVAAVKELLPMARAHATEFRRNAERFELREQTRRAHMSLSIPALSERAMARLMEIEAV
RSQGGDDAYKTAFALAAKDRSVVREIKAVSEALTARFGWSAFSAKADAIAERNIVERMPEDLTDEWRGKL
TRLFDAVRRFADEQHLAERRDRSKVVAGASADLSKEPGTEKIIMPPMFAAVTEFKVPIDDEARSRALASP
VYRQQRAALANAATTIWRDPAEVVGKIEELLQKGFAAERIGAAVTNNPAAYGALRGSDRLMDRMLTSGRE
RKEAVAAVPEAAARLRALGAAHLNALDAERQAITDERRRMAVAIPALSKAAEEALAHLTVEVSKDSRKLS
VSAASLDPGIGREFAAVSRALDERFGRNALVRGDKDIANVVPPAQRGAFAAMQERLKVLQQTVRLQSSEQ
IIVERRQQTANRSRGINL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1704 | GenBank | WP_012489500 |
| Name | traG_AMC78_RS24290_pRphaN161b |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70607.76 Da Isoelectric Point: 9.5676
>WP_012489500.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MGLRGKPHPSLLLVLVPIAVTTITIYIVGWRWPGLAAGMSGKIEHWFLRAAPVPPLLFGPLAGLLTVWAL
PLHRRRPVAMASLLYFLGVAAFYALREFGRLAPAVQAEVITWDRALSYLDMVAVIAAVAGFMAVAMSARI
SVVVPDEIKRARRGIFGDADWLPMTAAGKLFPPEGEIVVGERYRVDKEIVHALPFDANDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYSGPLICLDPSTEVAPMVAGHRARALKREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSGSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNISASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAK
VNRFVKPVL
MGLRGKPHPSLLLVLVPIAVTTITIYIVGWRWPGLAAGMSGKIEHWFLRAAPVPPLLFGPLAGLLTVWAL
PLHRRRPVAMASLLYFLGVAAFYALREFGRLAPAVQAEVITWDRALSYLDMVAVIAAVAGFMAVAMSARI
SVVVPDEIKRARRGIFGDADWLPMTAAGKLFPPEGEIVVGERYRVDKEIVHALPFDANDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYSGPLICLDPSTEVAPMVAGHRARALKREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSGSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNISASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAK
VNRFVKPVL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 157873..167460
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AMC78_RS34505 | 154367..154593 | - | 227 | Protein_139 | chaperonin GroEL | - |
| AMC78_RS24165 (AMC78_PB00123) | 154888..155808 | + | 921 | WP_029875459 | zincin-like metallopeptidase domain-containing protein | - |
| AMC78_RS24170 | 155919..156101 | - | 183 | WP_009993909 | hypothetical protein | - |
| AMC78_RS24175 (AMC78_PB00125) | 156863..157234 | - | 372 | WP_012489483 | hypothetical protein | - |
| AMC78_RS24180 (AMC78_PB00126) | 157333..157869 | + | 537 | WP_012489484 | hypothetical protein | - |
| AMC78_RS24185 (AMC78_PB00127) | 157873..158514 | + | 642 | WP_009991940 | transglycosylase SLT domain-containing protein | virB1 |
| AMC78_RS24190 (AMC78_PB00128) | 158529..158828 | + | 300 | WP_009991942 | TrbC/VirB2 family protein | virB2 |
| AMC78_RS24195 (AMC78_PB00129) | 158834..159172 | + | 339 | WP_010068249 | type IV secretion system protein VirB3 | virB3 |
| AMC78_RS24200 (AMC78_PB00130) | 159165..161531 | + | 2367 | WP_029875460 | VirB4 family type IV secretion system protein | virb4 |
| AMC78_RS24205 (AMC78_PB00131) | 161528..162229 | + | 702 | WP_009996884 | P-type DNA transfer protein VirB5 | virB5 |
| AMC78_RS24210 (AMC78_PB00132) | 162226..162459 | + | 234 | WP_064836762 | EexN family lipoprotein | - |
| AMC78_RS24215 (AMC78_PB00133) | 162462..163394 | + | 933 | WP_009996881 | type IV secretion system protein | virB6 |
| AMC78_RS34510 (AMC78_PB00134) | 163434..163715 | + | 282 | WP_012489488 | hypothetical protein | - |
| AMC78_RS24220 (AMC78_PB00135) | 163717..164388 | + | 672 | WP_029875461 | virB8 family protein | virB8 |
| AMC78_RS24225 (AMC78_PB00136) | 164385..165242 | + | 858 | WP_009996879 | P-type conjugative transfer protein VirB9 | virB9 |
| AMC78_RS24230 (AMC78_PB00137) | 165251..166423 | + | 1173 | WP_012489490 | type IV secretion system protein VirB10 | virB10 |
| AMC78_RS24235 (AMC78_PB00138) | 166432..167460 | + | 1029 | WP_012489491 | P-type DNA transfer ATPase VirB11 | virB11 |
| AMC78_RS24240 (AMC78_PB00139) | 167504..168076 | - | 573 | WP_010067915 | PIN domain-containing protein | - |
| AMC78_RS24245 (AMC78_PB00140) | 168066..168512 | - | 447 | WP_029875463 | hypothetical protein | - |
| AMC78_RS24250 (AMC78_PB00142) | 169025..170215 | - | 1191 | WP_012489493 | DUF1173 domain-containing protein | - |
| AMC78_RS34515 | 170526..170785 | + | 260 | Protein_159 | toprim domain-containing protein | - |
| AMC78_RS24255 (AMC78_PB00144) | 171173..171439 | + | 267 | WP_004676093 | hypothetical protein | - |
| AMC78_RS24260 (AMC78_PB00145) | 171436..171834 | + | 399 | WP_008536014 | type II toxin-antitoxin system VapC family toxin | - |
Host bacterium
| ID | 3220 | GenBank | NZ_CP013587 |
| Plasmid name | pRphaN161b | Incompatibility group | - |
| Plasmid size | 395855 bp | Coordinate of oriT [Strand] | 177632..177660 [+] |
| Host baterium | Rhizobium phaseoli strain N161 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | hsiC1/vipB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nifX, nifN, nifE, nifK, nifD, nifH, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, nifT, nodD, nodJ, nodI, nodS, nodC, fixO, fixN, mLTONO_5203, nodA |
| Anti-CRISPR | - |