Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102769 |
| Name | oriT_pRphaN671e |
| Organism | Rhizobium phaseoli strain N671 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013579 (1069212..1069240 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRphaN671e
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file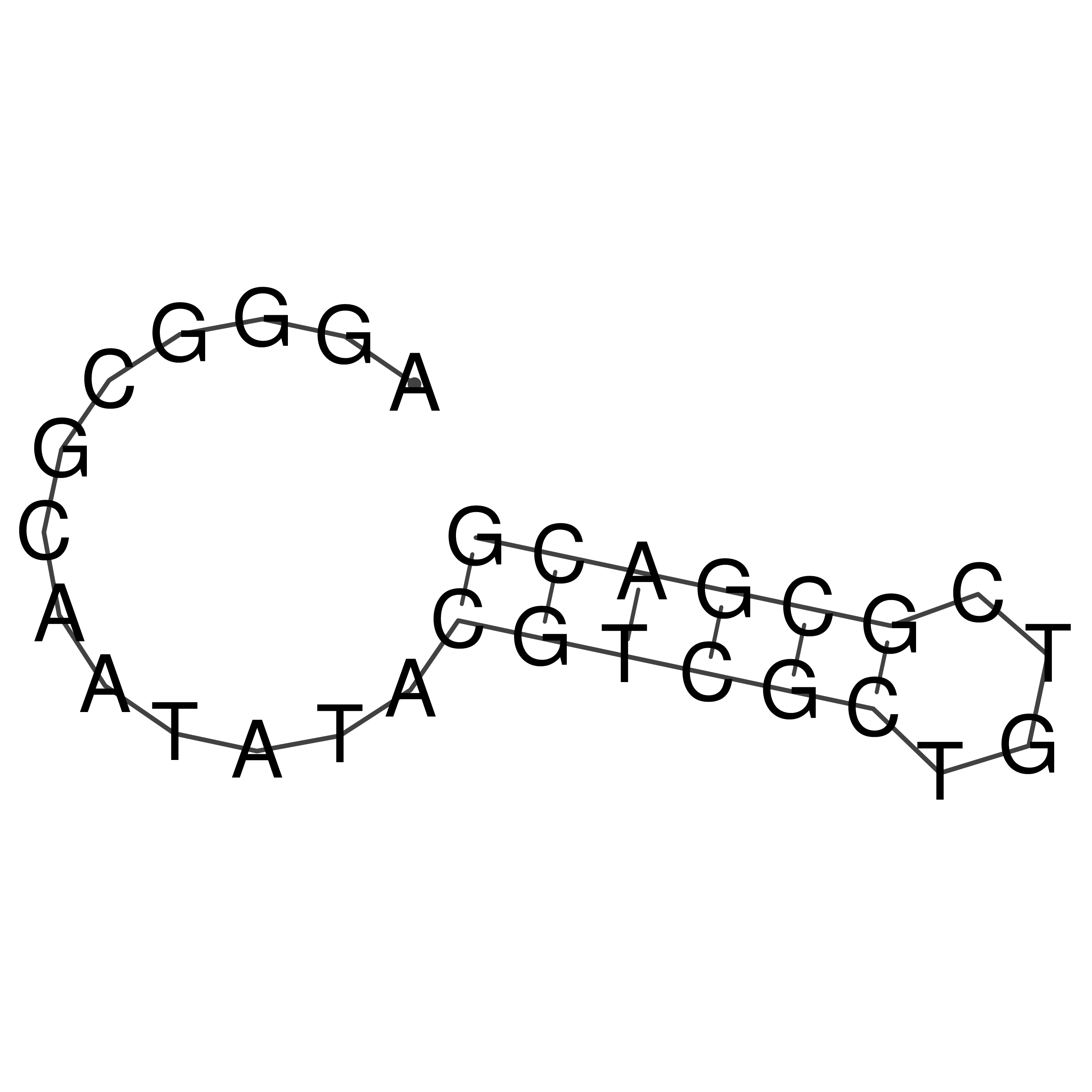
Relaxase
| ID | 2042 | GenBank | WP_037146755 |
| Name | traA_AMC80_RS32515_pRphaN671e |
UniProt ID | _ |
| Length | 1536 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1536 a.a. Molecular weight: 170772.21 Da Isoelectric Point: 7.4234
>WP_037146755.1 Ti-type conjugative transfer relaxase TraA [Rhizobium phaseoli]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGAASELMHEELALPDQIPAWLKMAMDG
RSIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEHSKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGVAQKHLGPEKAALARKGRELFFAPADLARRQGMADRLLAEPELLLKQ
LGNERSTFDEKDIAKALHRCVDDPVEFANIRARLMASDELVMLKPQEMDADTGKVSEPAVFTTRDMLRIE
YHMAESARRLSNRSGYAVSRKHVAAAIERIESEDPKNLFHLDAEQVDAIRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAEIKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VEKGLDAYAQQGHLVEAGTREEAINRIVADWTEARKQAIDRSTSEGRDGHLRGDELLVLAHTNADVHKLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPLMSVLLDNGRNVVFSEDSYRNVDHGYASTIHKSQGATVDRTFVLATGMMDRHLTYVSMTRHRDRADL
YAVKEDFQVRSERGRRLRADHAAGVTGELVEAGEAKFRPDDEDADDSPYADIRTDDGTVHRLWGVSLPKA
LEEAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYEEREVGRNVWTAKQIETAEARQERIERESHRP
DVFSRLVERLSRSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGLAIERERRVAYNEERSETMIGAISSEERYLIPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASDTGTEPRKLADDLAVAPDLLGRLYGSDLIVDGRAARDERNVAMA
TVKELLPLARAHATEFRRNATRFERREQTRRNYMVLSIPALSERAVARLTEIEAVRSRGGDDAYKTASAL
AAQDRTVVQEIKAVSEAITARFGSSAFTAKADAIAQRNMAERMLEELTSVRGEKLIRLFEAVRRFAEEQH
LAERQDRSKIVAGASADPQNQNVPMLAAVTEFKTPIDEEARSRVLAGPLYRQQRAALAAVASTIWRDPAG
AVDKIEELLVKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVPEAAARLRSLGSAY
VNVLDAERQAIAEERRRMAVAIPGLSKSAEEVLMRLTTEAKNNGRTRNAAAASLDPSIHREFEAVSRALD
ERFGHRAIIRGEKDLFNRMPLAQRRAFEVMWERFKVLQQTVRWESSEKIVSELRNRTINRGRGIDL
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGAASELMHEELALPDQIPAWLKMAMDG
RSIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEHSKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGVAQKHLGPEKAALARKGRELFFAPADLARRQGMADRLLAEPELLLKQ
LGNERSTFDEKDIAKALHRCVDDPVEFANIRARLMASDELVMLKPQEMDADTGKVSEPAVFTTRDMLRIE
YHMAESARRLSNRSGYAVSRKHVAAAIERIESEDPKNLFHLDAEQVDAIRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAEIKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VEKGLDAYAQQGHLVEAGTREEAINRIVADWTEARKQAIDRSTSEGRDGHLRGDELLVLAHTNADVHKLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPLMSVLLDNGRNVVFSEDSYRNVDHGYASTIHKSQGATVDRTFVLATGMMDRHLTYVSMTRHRDRADL
YAVKEDFQVRSERGRRLRADHAAGVTGELVEAGEAKFRPDDEDADDSPYADIRTDDGTVHRLWGVSLPKA
LEEAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYEEREVGRNVWTAKQIETAEARQERIERESHRP
DVFSRLVERLSRSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGLAIERERRVAYNEERSETMIGAISSEERYLIPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASDTGTEPRKLADDLAVAPDLLGRLYGSDLIVDGRAARDERNVAMA
TVKELLPLARAHATEFRRNATRFERREQTRRNYMVLSIPALSERAVARLTEIEAVRSRGGDDAYKTASAL
AAQDRTVVQEIKAVSEAITARFGSSAFTAKADAIAQRNMAERMLEELTSVRGEKLIRLFEAVRRFAEEQH
LAERQDRSKIVAGASADPQNQNVPMLAAVTEFKTPIDEEARSRVLAGPLYRQQRAALAAVASTIWRDPAG
AVDKIEELLVKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVPEAAARLRSLGSAY
VNVLDAERQAIAEERRRMAVAIPGLSKSAEEVLMRLTTEAKNNGRTRNAAAASLDPSIHREFEAVSRALD
ERFGHRAIIRGEKDLFNRMPLAQRRAFEVMWERFKVLQQTVRWESSEKIVSELRNRTINRGRGIDL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1696 | GenBank | WP_037146751 |
| Name | traG_AMC80_RS32530_pRphaN671e |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70465.33 Da Isoelectric Point: 9.2555
>WP_037146751.1 Ti-type conjugative transfer system protein TraG [Rhizobium phaseoli]
MIARGKPHPSLLFVFAPVVVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVFASYAMREFARLGPSVQTGVITWDRALSYLDMVAVLGAVAGFLAVAVSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDANDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHALGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQVIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
MIARGKPHPSLLFVFAPVVVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVFASYAMREFARLGPSVQTGVITWDRALSYLDMVAVLGAVAGFLAVAVSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDANDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHALGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQVIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1080515..1090070
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AMC80_RS32560 | 1076086..1076268 | + | 183 | WP_037146733 | hypothetical protein | - |
| AMC80_RS32565 (AMC80_PE01009) | 1076401..1077441 | - | 1041 | WP_037146729 | toprim domain-containing protein | - |
| AMC80_RS32570 (AMC80_PE01010) | 1077804..1078994 | + | 1191 | WP_037146725 | DUF1173 domain-containing protein | - |
| AMC80_RS32575 (AMC80_PE01011) | 1079437..1080030 | + | 594 | WP_037146723 | DUF433 domain-containing protein | - |
| AMC80_RS32580 (AMC80_PE01012) | 1080146..1080496 | + | 351 | WP_237353383 | type II toxin-antitoxin system VapC family toxin | - |
| AMC80_RS32585 (AMC80_PE01013) | 1080515..1081510 | - | 996 | WP_064832842 | P-type DNA transfer ATPase VirB11 | virB11 |
| AMC80_RS32590 (AMC80_PE01014) | 1081519..1082694 | - | 1176 | WP_016735275 | type IV secretion system protein VirB10 | virB10 |
| AMC80_RS32595 (AMC80_PE01015) | 1082703..1083557 | - | 855 | WP_037146719 | P-type conjugative transfer protein VirB9 | virB9 |
| AMC80_RS32600 (AMC80_PE01016) | 1083554..1084225 | - | 672 | WP_037146717 | virB8 family protein | virB8 |
| AMC80_RS35950 (AMC80_PE01017) | 1084219..1084506 | - | 288 | WP_080712055 | hypothetical protein | - |
| AMC80_RS32605 (AMC80_PE01018) | 1084546..1085478 | - | 933 | WP_037146716 | type IV secretion system protein | virB6 |
| AMC80_RS32610 (AMC80_PE01019) | 1085481..1085714 | - | 234 | WP_016735278 | EexN family lipoprotein | - |
| AMC80_RS32615 (AMC80_PE01020) | 1085711..1086412 | - | 702 | WP_016735279 | P-type DNA transfer protein VirB5 | virB5 |
| AMC80_RS32620 (AMC80_PE01021) | 1086409..1088775 | - | 2367 | WP_064826470 | VirB4 family type IV secretion system protein | virb4 |
| AMC80_RS32625 (AMC80_PE01022) | 1088768..1089106 | - | 339 | WP_016735282 | type IV secretion system protein VirB3 | virB3 |
| AMC80_RS32630 (AMC80_PE01023) | 1089112..1089411 | - | 300 | WP_016735283 | TrbC/VirB2 family protein | virB2 |
| AMC80_RS32635 (AMC80_PE01024) | 1089408..1090070 | - | 663 | WP_037147233 | transglycosylase SLT domain-containing protein | virB1 |
| AMC80_RS32640 (AMC80_PE01025) | 1090074..1090610 | - | 537 | WP_016735285 | hypothetical protein | - |
| AMC80_RS32645 (AMC80_PE01026) | 1090709..1091080 | + | 372 | WP_016735286 | hypothetical protein | - |
| AMC80_RS32650 (AMC80_PE01028) | 1091389..1092867 | - | 1479 | WP_189636755 | group II intron reverse transcriptase/maturase | - |
| AMC80_RS32655 (AMC80_PE01030) | 1093346..1094536 | - | 1191 | WP_037147230 | hypothetical protein | - |
Host bacterium
| ID | 3212 | GenBank | NZ_CP013579 |
| Plasmid name | pRphaN671e | Incompatibility group | - |
| Plasmid size | 1189373 bp | Coordinate of oriT [Strand] | 1069212..1069240 [-] |
| Host baterium | Rhizobium phaseoli strain N671 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | hupE2, actP |
| Degradation gene | LinG, dxnH |
| Symbiosis gene | noeL, fixB, fixA |
| Anti-CRISPR | AcrIIA8 |