Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102758 |
| Name | oriT_pRphaR744d |
| Organism | Rhizobium phaseoli strain R744 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013526 (1083009..1083037 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRphaR744d
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file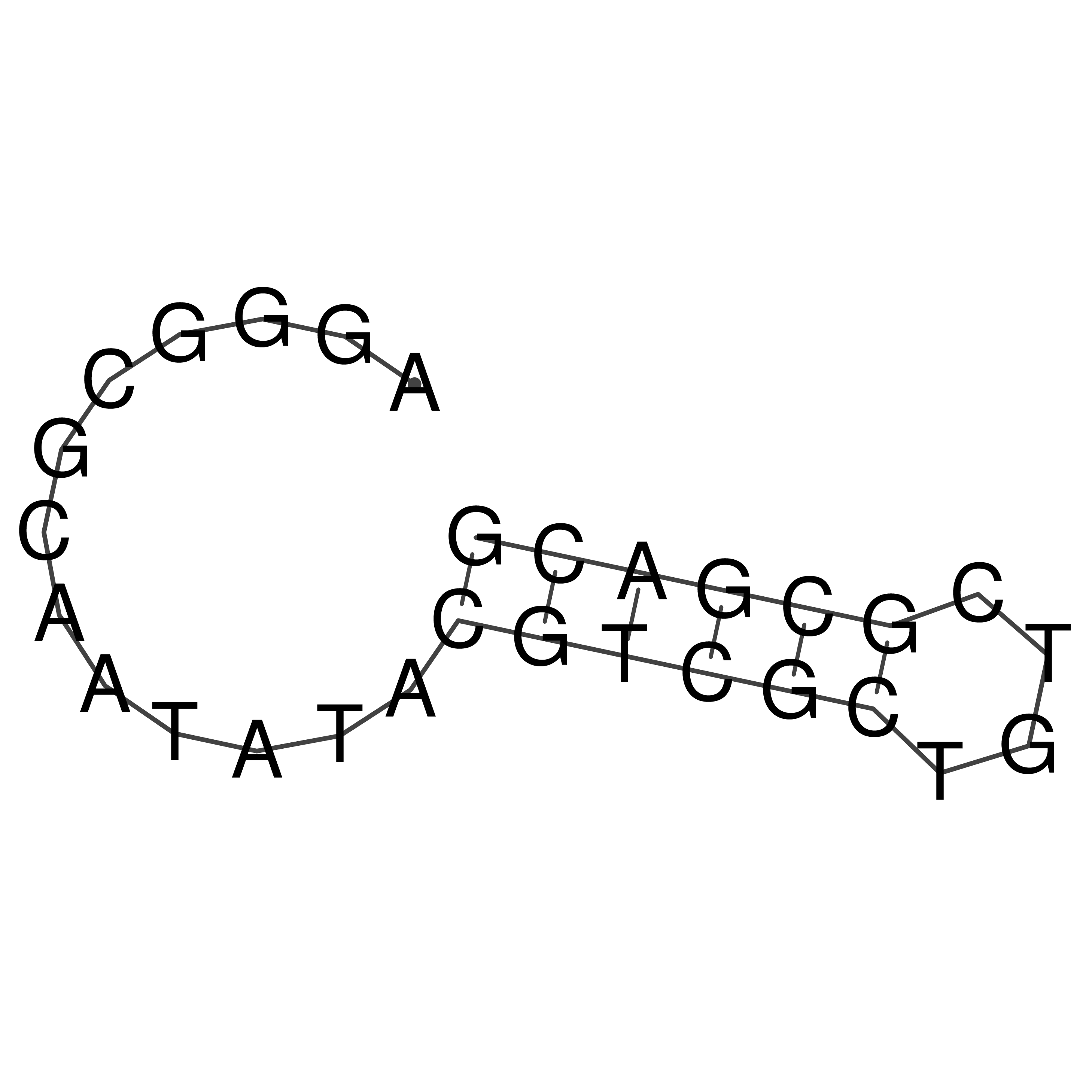
Relaxase
| ID | 2032 | GenBank | WP_064844904 |
| Name | traA_AMC90_RS31020_pRphaR744d |
UniProt ID | _ |
| Length | 1543 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1543 a.a. Molecular weight: 170693.24 Da Isoelectric Point: 7.0315
>WP_064844904.1 Ti-type conjugative transfer relaxase TraA [Rhizobium phaseoli]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELMHEELALPEQIPTWLREAIAG
QPIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALIREFVSGNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEDGKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARQGRELFFAPADLARRQEMADRLLAEPELLLKH
LGNERSTFDEKDIAKALHRYVDDPVEFANIRARLMASDELVMLKPQEMDAGTGKVSEPAVFTTRDMLRIE
YHMAESARRLSKLSGYAVLRKHVAAAIERIESDEPKNLFRLDAEQVDAIRHVTGDSGIAAIVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELAWANGRETLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAEIKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VEKGLDAYAQQGHLVEAGTREEAINRIVADWTEARKQAIDRSTSEGRNGHLRGDELLVLAHTNADVKRLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPLLSVLLDNGRNVVFSEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDRHLTYVSMTRHRDRADL
YAAKEDFAARPEWGHKARVDHAAGVTGELVEAGEAKFRPDDEDADDSPYADIRTDDGSVHRLWGVSLPKA
LEQAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYEEREVGRNVWTAKQIETAEARQERIERESHRP
DVFSRLVERLARSGSKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGFAIERERRVAYNEERSETMIGAISSEERYLMPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASDTGTEPRGLADNLAVAPDRLGRLRGSELIVEGRAARDERDVAMA
ALEELLPLVRGHATEFRRNAERFELREQTRRASMSLSVPALSEQAMARLMEIEAVRTQGGDDAYKTAFAL
AAEDRSVVQEVKAVSEALTARFGWSAFTAKADAMAERNMVERMPEDLSAGRREELIRLFKAVKRFAEEQH
LAERRDRSKIVAAASAVPGAAPGKQTVTVLPMLAAVTEFKTSVDDEARSRAIAAPLYRQQRAALAEAARM
IWRDPAATVDNIESLLEKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVPEAAARL
RSLGSAYVNVLDAERQAIAEERRRMAVAIPGLSKAAEGVLMRLTTEVKNNGRTRNAAAGSLDPSIHREFD
AVSRALDERFGRSAIARGEKDLLNRMPPAQRAAFAAMQEKLKVLQQTVRWESSEKIVSELRSRTVNRGRG
IEL
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELMHEELALPEQIPTWLREAIAG
QPIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALIREFVSGNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEDGKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARQGRELFFAPADLARRQEMADRLLAEPELLLKH
LGNERSTFDEKDIAKALHRYVDDPVEFANIRARLMASDELVMLKPQEMDAGTGKVSEPAVFTTRDMLRIE
YHMAESARRLSKLSGYAVLRKHVAAAIERIESDEPKNLFRLDAEQVDAIRHVTGDSGIAAIVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELAWANGRETLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAEIKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VEKGLDAYAQQGHLVEAGTREEAINRIVADWTEARKQAIDRSTSEGRNGHLRGDELLVLAHTNADVKRLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPLLSVLLDNGRNVVFSEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDRHLTYVSMTRHRDRADL
YAAKEDFAARPEWGHKARVDHAAGVTGELVEAGEAKFRPDDEDADDSPYADIRTDDGSVHRLWGVSLPKA
LEQAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYEEREVGRNVWTAKQIETAEARQERIERESHRP
DVFSRLVERLARSGSKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGFAIERERRVAYNEERSETMIGAISSEERYLMPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASDTGTEPRGLADNLAVAPDRLGRLRGSELIVEGRAARDERDVAMA
ALEELLPLVRGHATEFRRNAERFELREQTRRASMSLSVPALSEQAMARLMEIEAVRTQGGDDAYKTAFAL
AAEDRSVVQEVKAVSEALTARFGWSAFTAKADAMAERNMVERMPEDLSAGRREELIRLFKAVKRFAEEQH
LAERRDRSKIVAAASAVPGAAPGKQTVTVLPMLAAVTEFKTSVDDEARSRAIAAPLYRQQRAALAEAARM
IWRDPAATVDNIESLLEKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVPEAAARL
RSLGSAYVNVLDAERQAIAEERRRMAVAIPGLSKAAEGVLMRLTTEVKNNGRTRNAAAGSLDPSIHREFD
AVSRALDERFGRSAIARGEKDLLNRMPPAQRAAFAAMQEKLKVLQQTVRWESSEKIVSELRSRTVNRGRG
IEL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1682 | GenBank | WP_064844908 |
| Name | traG_AMC90_RS31035_pRphaR744d |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70315.18 Da Isoelectric Point: 9.1203
>WP_064844908.1 Ti-type conjugative transfer system protein TraG [Rhizobium phaseoli]
MIARGKPHPSLLFVLAPVLVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVSAFYALREFARLGPSVQTGVITWDGALSYLDMVAVLGAVAGFLAVAVSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDASDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHALGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
MIARGKPHPSLLFVLAPVLVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVSAFYALREFARLGPSVQTGVITWDGALSYLDMVAVLGAVAGFLAVAVSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDASDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHALGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1094884..1104440
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AMC90_RS31075 (AMC90_PD01012) | 1090466..1090648 | + | 183 | WP_016735269 | hypothetical protein | - |
| AMC90_RS31080 (AMC90_PD01013) | 1090780..1091817 | - | 1038 | WP_064844920 | toprim domain-containing protein | - |
| AMC90_RS31085 (AMC90_PD01014) | 1092176..1093366 | + | 1191 | WP_064844922 | DUF1173 domain-containing protein | - |
| AMC90_RS31090 (AMC90_PD01015) | 1093809..1094402 | + | 594 | WP_064844924 | DUF433 domain-containing protein | - |
| AMC90_RS31095 (AMC90_PD01016) | 1094512..1094865 | + | 354 | WP_237359419 | type II toxin-antitoxin system VapC family toxin | - |
| AMC90_RS31100 (AMC90_PD01017) | 1094884..1095879 | - | 996 | WP_064829760 | P-type DNA transfer ATPase VirB11 | virB11 |
| AMC90_RS31105 (AMC90_PD01018) | 1095888..1097063 | - | 1176 | WP_064844926 | type IV secretion system protein VirB10 | virB10 |
| AMC90_RS31110 (AMC90_PD01019) | 1097072..1097926 | - | 855 | WP_064844928 | P-type conjugative transfer protein VirB9 | virB9 |
| AMC90_RS31115 (AMC90_PD01020) | 1097923..1098594 | - | 672 | WP_064844930 | virB8 family protein | virB8 |
| AMC90_RS33985 (AMC90_PD01021) | 1098588..1098875 | - | 288 | WP_012490329 | hypothetical protein | - |
| AMC90_RS31120 (AMC90_PD01022) | 1098916..1099848 | - | 933 | WP_012490330 | type IV secretion system protein | virB6 |
| AMC90_RS31125 (AMC90_PD01023) | 1099851..1100084 | - | 234 | WP_016735278 | EexN family lipoprotein | - |
| AMC90_RS31130 (AMC90_PD01024) | 1100081..1100782 | - | 702 | WP_064844932 | P-type DNA transfer protein VirB5 | virB5 |
| AMC90_RS31135 (AMC90_PD01025) | 1100779..1103145 | - | 2367 | WP_064844934 | VirB4 family type IV secretion system protein | virb4 |
| AMC90_RS31140 (AMC90_PD01026) | 1103138..1103476 | - | 339 | WP_064837799 | type IV secretion system protein VirB3 | virB3 |
| AMC90_RS31145 (AMC90_PD01027) | 1103482..1103781 | - | 300 | WP_064844936 | TrbC/VirB2 family protein | virB2 |
| AMC90_RS31150 (AMC90_PD01028) | 1103778..1104440 | - | 663 | WP_064844938 | transglycosylase SLT domain-containing protein | virB1 |
| AMC90_RS31155 (AMC90_PD01029) | 1104444..1104980 | - | 537 | WP_064844940 | hypothetical protein | - |
| AMC90_RS31160 (AMC90_PD01030) | 1105079..1105450 | + | 372 | WP_004679908 | hypothetical protein | - |
| AMC90_RS31165 (AMC90_PD01031) | 1105804..1106994 | - | 1191 | WP_037147230 | hypothetical protein | - |
| AMC90_RS31170 (AMC90_PD01032) | 1107068..1107724 | - | 657 | WP_064844942 | hypothetical protein | - |
Host bacterium
| ID | 3201 | GenBank | NZ_CP013526 |
| Plasmid name | pRphaR744d | Incompatibility group | - |
| Plasmid size | 1195489 bp | Coordinate of oriT [Strand] | 1083009..1083037 [-] |
| Host baterium | Rhizobium phaseoli strain R744 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | hupE2, actP |
| Degradation gene | - |
| Symbiosis gene | noeL, fixB, fixA |
| Anti-CRISPR | AcrIIA8 |