Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102754 |
| Name | oriT_pRspN741e |
| Organism | Rhizobium sp. N741 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013600 (709453..709481 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRspN741e
AGGGCGCAATATACGTCGCTGCCGCGACG
AGGGCGCAATATACGTCGCTGCCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file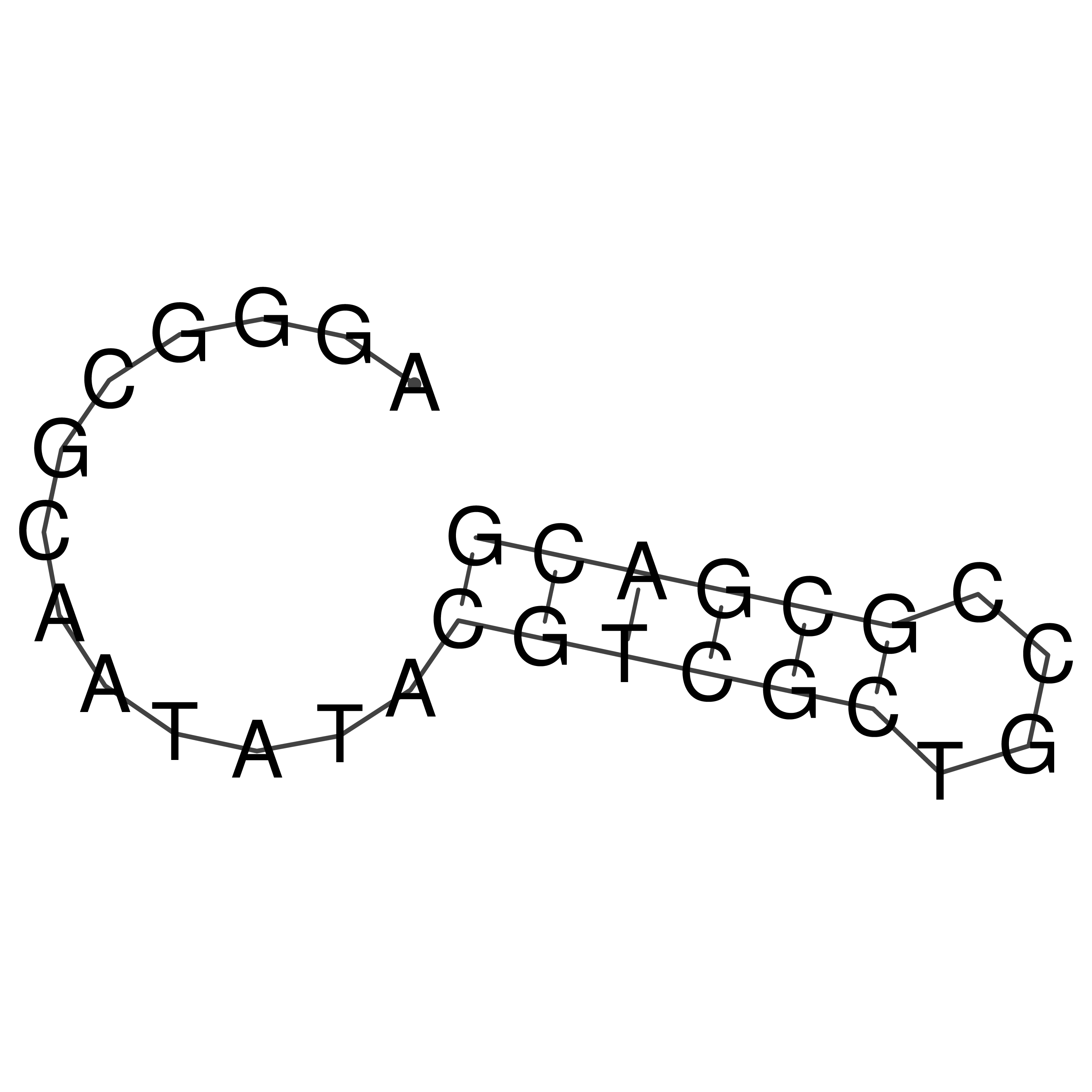
Relaxase
| ID | 2028 | GenBank | WP_064813248 |
| Name | traA_AMK03_RS30975_pRspN741e |
UniProt ID | _ |
| Length | 1553 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1553 a.a. Molecular weight: 171806.03 Da Isoelectric Point: 6.7399
>WP_064813248.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGSRELVHEELALPEQVPGWLRAAIDG
QSVAGASEALWNAVDAFETRADGQLARELIIALPEELTRAENIALVREFVHDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTEDGFGPKKVPVLGEDDKPLRVVTPDRPNGKIVYKVWAGDKETMKGWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELYFAPADLARRHEMADRLLAEPELLLKQ
LGNERSTFDEKDIAKALHRYVDDPSDFANIRARLMASDELVMLKPQEFDGETREVAAPAVFTTREMLRIE
YDMARSARVLSEQAGFGVSARQVDTAIERVETGDPAKPFKLDAEQVDAIRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLTAWELAWANGRDTLHRGDVLVIDEAGMVA
SQQMARVLDIVEEASAKVVLVGDAMQLQPIQAGAAFRAISDRIGFAELAGVRRQREEWAREASRLFARGE
VEKGLEAYAQHGHLAEAGTRAETIDRIVGDWSEARREAIGRSVSEGRDGRLRGDELLVLAHTNDDVRKLN
EALRAVMAGQRALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARSYGPQYVKNGMLGTVISTGDKR
GEALMSVRLDNGHKVVFSEDSYRNVDHGYAATIHKAQGATVERTLVLATGMMDQHLTYVAMTRHRDRVDL
YAASEDFEPRPEWGRKPRVDHAAGVTGELVETGEAKFRPDDEDADDSPYADVRTDDGTVHRLWGVSLPKA
LEEGGLSEGDTVTLRKDGVERVKVQIAVIDEETGQKRYEEREVDRNVWTARQIETAEARQDRIERESHRP
DLFKPLVERLSRSGAKTTTLDFESEAGYRAYAEDFARRRGFEYLSLAAAEIEGDISRRFAWIAAKREQQE
KLWERASMALGFAIERERRVSYSEARTEARPVPSANAGKAHYLILPTTEFVRSIDEDARLAQLSSPVWKD
RETLLRPVLERIYRDPDGALVALNALASDAGTEPRRLADDLAVSPDRLGRLRGSELIVDGRAARDERNVA
LAAVTELLPMARAHASEFRRNAERFELNEQARRAHMSLSIPALSEGAMARLMEIEAVRHRGGDDAYKTAF
ALAAKDRTVVQEIKAVSEALSARFGWSAFTAKADAVAERNMAERMPEALMAGEREKLVHLFEAVKRFAEA
QHLAERHDRSKIVAAASAVADQEQGMQPGMKSGKENVTVLPMLAAVTEFKTAVDDEARSRVLSAPFYRQQ
RAALANAARTIWRDPAGAVGKIEELIVKGFAAERIAAAVSNDPAAYGALRGSGRLMDKMLASGRERKEAV
QAVPEAAARLRSLGSAYANIFDAERQAIKAERERMAVAIPGLSKVAEEALVRLTAEARKNDRKRGIATAP
LDANIRREFDEISKALDQRFGRNAIIRGDKDLINHVPEPQRQAFVAMQEKLKILQQTVRAESSQQILSER
RQRGFNRGRGIDI
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGSRELVHEELALPEQVPGWLRAAIDG
QSVAGASEALWNAVDAFETRADGQLARELIIALPEELTRAENIALVREFVHDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTEDGFGPKKVPVLGEDDKPLRVVTPDRPNGKIVYKVWAGDKETMKGWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELYFAPADLARRHEMADRLLAEPELLLKQ
LGNERSTFDEKDIAKALHRYVDDPSDFANIRARLMASDELVMLKPQEFDGETREVAAPAVFTTREMLRIE
YDMARSARVLSEQAGFGVSARQVDTAIERVETGDPAKPFKLDAEQVDAIRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLTAWELAWANGRDTLHRGDVLVIDEAGMVA
SQQMARVLDIVEEASAKVVLVGDAMQLQPIQAGAAFRAISDRIGFAELAGVRRQREEWAREASRLFARGE
VEKGLEAYAQHGHLAEAGTRAETIDRIVGDWSEARREAIGRSVSEGRDGRLRGDELLVLAHTNDDVRKLN
EALRAVMAGQRALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARSYGPQYVKNGMLGTVISTGDKR
GEALMSVRLDNGHKVVFSEDSYRNVDHGYAATIHKAQGATVERTLVLATGMMDQHLTYVAMTRHRDRVDL
YAASEDFEPRPEWGRKPRVDHAAGVTGELVETGEAKFRPDDEDADDSPYADVRTDDGTVHRLWGVSLPKA
LEEGGLSEGDTVTLRKDGVERVKVQIAVIDEETGQKRYEEREVDRNVWTARQIETAEARQDRIERESHRP
DLFKPLVERLSRSGAKTTTLDFESEAGYRAYAEDFARRRGFEYLSLAAAEIEGDISRRFAWIAAKREQQE
KLWERASMALGFAIERERRVSYSEARTEARPVPSANAGKAHYLILPTTEFVRSIDEDARLAQLSSPVWKD
RETLLRPVLERIYRDPDGALVALNALASDAGTEPRRLADDLAVSPDRLGRLRGSELIVDGRAARDERNVA
LAAVTELLPMARAHASEFRRNAERFELNEQARRAHMSLSIPALSEGAMARLMEIEAVRHRGGDDAYKTAF
ALAAKDRTVVQEIKAVSEALSARFGWSAFTAKADAVAERNMAERMPEALMAGEREKLVHLFEAVKRFAEA
QHLAERHDRSKIVAAASAVADQEQGMQPGMKSGKENVTVLPMLAAVTEFKTAVDDEARSRVLSAPFYRQQ
RAALANAARTIWRDPAGAVGKIEELIVKGFAAERIAAAVSNDPAAYGALRGSGRLMDKMLASGRERKEAV
QAVPEAAARLRSLGSAYANIFDAERQAIKAERERMAVAIPGLSKVAEEALVRLTAEARKNDRKRGIATAP
LDANIRREFDEISKALDQRFGRNAIIRGDKDLINHVPEPQRQAFVAMQEKLKILQQTVRAESSQQILSER
RQRGFNRGRGIDI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1678 | GenBank | WP_064813078 |
| Name | traG_AMK03_RS30990_pRspN741e |
UniProt ID | _ |
| Length | 638 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 638 a.a. Molecular weight: 70312.12 Da Isoelectric Point: 9.0374
>WP_064813078.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTSKKRLHPSLLCVLVPVVVTAISAYVVNWRWPILAHGLTGTTEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMASLACFLIMAGCYALREYSRLAPFVQAGAVSLNGALSYLDLVAVIGAVMGFMAVAVAARI
SVVVPEAVKRVKGGAFGDADWLSMSAAAKLFPPNGEIVVGERYRVDKEIVHQLPFDPNDHSTWGQGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVTEHRTQVLGREVMVLDPN
NPINGFNVLDGIEHSRRKEEEIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSKSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GDAFRPSDIVAGRKDVFLNIPASILRSYPGIARVIIGSLVNAMVQADGDFSRRALFILDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEVTVEVQG
RSRNIGWDAKGGTRRSESVNVQRRPLILPHEITQAMRKDEQIIIVQGHNPIRCGRAIYFRRKEMDRAAKA
NRFVKPEQ
MTSKKRLHPSLLCVLVPVVVTAISAYVVNWRWPILAHGLTGTTEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMASLACFLIMAGCYALREYSRLAPFVQAGAVSLNGALSYLDLVAVIGAVMGFMAVAVAARI
SVVVPEAVKRVKGGAFGDADWLSMSAAAKLFPPNGEIVVGERYRVDKEIVHQLPFDPNDHSTWGQGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVTEHRTQVLGREVMVLDPN
NPINGFNVLDGIEHSRRKEEEIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSKSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GDAFRPSDIVAGRKDVFLNIPASILRSYPGIARVIIGSLVNAMVQADGDFSRRALFILDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEVTVEVQG
RSRNIGWDAKGGTRRSESVNVQRRPLILPHEITQAMRKDEQIIIVQGHNPIRCGRAIYFRRKEMDRAAKA
NRFVKPEQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 721629..730559
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AMK03_RS31030 (AMK03_PE00658) | 716635..717576 | - | 942 | WP_064813094 | DUF2493 domain-containing protein | - |
| AMK03_RS31035 (AMK03_PE00659) | 717900..718934 | - | 1035 | WP_064813096 | toprim domain-containing protein | - |
| AMK03_RS31045 | 719351..719551 | + | 201 | WP_145924266 | hypothetical protein | - |
| AMK03_RS31050 (AMK03_PE00661) | 719663..720853 | + | 1191 | WP_064813102 | DUF1173 domain-containing protein | - |
| AMK03_RS31055 (AMK03_PE00662) | 720893..721291 | - | 399 | WP_064813104 | type II toxin-antitoxin system VapC family toxin | - |
| AMK03_RS31060 (AMK03_PE00663) | 721288..721539 | - | 252 | WP_064813105 | AbrB/MazE/SpoVT family DNA-binding domain-containing protein | - |
| AMK03_RS31065 (AMK03_PE00664) | 721629..722651 | - | 1023 | WP_064813107 | P-type DNA transfer ATPase VirB11 | virB11 |
| AMK03_RS31070 (AMK03_PE00665) | 722660..723835 | - | 1176 | WP_064813109 | type IV secretion system protein VirB10 | virB10 |
| AMK03_RS31075 (AMK03_PE00666) | 723844..724701 | - | 858 | WP_064813111 | P-type conjugative transfer protein VirB9 | virB9 |
| AMK03_RS31080 (AMK03_PE00667) | 724698..725369 | - | 672 | WP_064813251 | virB8 family protein | virB8 |
| AMK03_RS34220 | 725363..725656 | - | 294 | WP_082240409 | hypothetical protein | - |
| AMK03_RS31085 (AMK03_PE00668) | 725694..726626 | - | 933 | WP_064813113 | type IV secretion system protein | virB6 |
| AMK03_RS31090 (AMK03_PE00669) | 726629..726862 | - | 234 | WP_064813115 | EexN family lipoprotein | - |
| AMK03_RS31095 (AMK03_PE00670) | 726859..727557 | - | 699 | WP_064813117 | P-type DNA transfer protein VirB5 | virB5 |
| AMK03_RS31100 (AMK03_PE00671) | 727557..729923 | - | 2367 | WP_097585286 | VirB4 family type IV secretion system protein | virb4 |
| AMK03_RS31105 (AMK03_PE00672) | 729916..730254 | - | 339 | WP_064813121 | type IV secretion system protein VirB3 | virB3 |
| AMK03_RS31110 (AMK03_PE00673) | 730260..730559 | - | 300 | WP_064813123 | TrbC/VirB2 family protein | virB2 |
| AMK03_RS31115 (AMK03_PE00674) | 730556..731222 | - | 667 | Protein_693 | lytic transglycosylase domain-containing protein | - |
| AMK03_RS32550 (AMK03_PE00675) | 731225..731755 | - | 531 | WP_082240410 | hypothetical protein | - |
| AMK03_RS31120 (AMK03_PE00676) | 731857..732228 | + | 372 | WP_064813253 | hypothetical protein | - |
| AMK03_RS31125 (AMK03_PE00677) | 732617..733807 | - | 1191 | WP_064813127 | hypothetical protein | - |
Host bacterium
| ID | 3197 | GenBank | NZ_CP013600 |
| Plasmid name | pRspN741e | Incompatibility group | - |
| Plasmid size | 752910 bp | Coordinate of oriT [Strand] | 709453..709481 [-] |
| Host baterium | Rhizobium sp. N741 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | hupE2 |
| Degradation gene | - |
| Symbiosis gene | fixB, fixA |
| Anti-CRISPR | - |